Fig. 2.
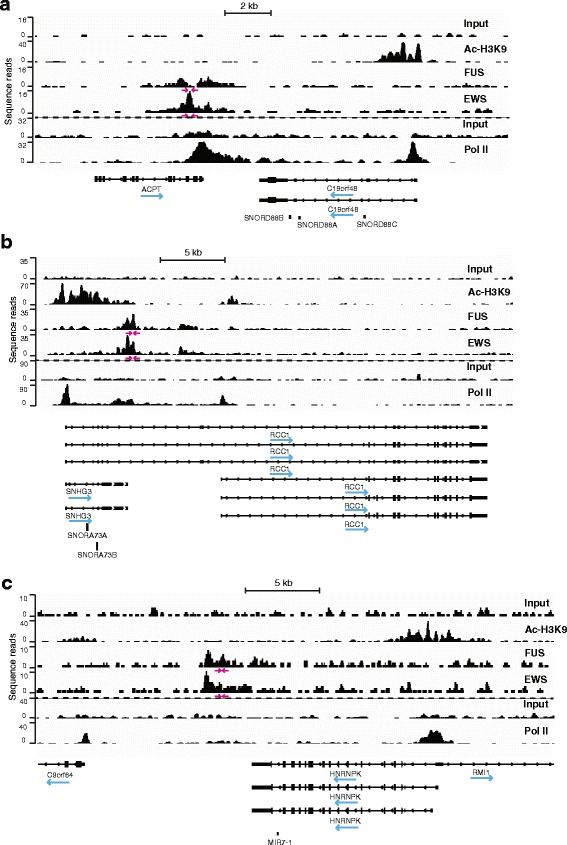
Graphical illustrations of representative ChIP-seq results. ChIP-seq sequence reads for input, FUS, EWS, Ac-H3K9 and RNAPII were aligned to representative genes by Integrative Genomics Viewer 2.0. The number of sequence reads is presented on the y-axis within each figure. Transcripts from the UCSC hg19 genomic database are shown in the bottom section of each figure. a ChIP-seq reads aligned to the neighboring ACPT and C19orf48 genes. b ChIP-seq reads aligned to the overlapping RCC1 and SNHG3 genes with additional inclusion of SNORA73A and SNORA73B genes. We note the presence of major FUS and EWS peaks corresponding to the position of the annotated poly(A)-signal of SNHG3 as well as minor FUS and EWS peaks 4 kb further 3’ end positioned. c ChIP-seq reads aligned to the HNRNPK gene with additional inclusion of miR-7-1. Dashed lines separate the paired input, ac-H3K9, FUS and EWS ChIP seq dataset from the paired input and RNAPII ChIP-seq dataset. Red arrows show positions of primers used for qPCR based verification of ChIP results. Blue arrows under gene names show direction of transcription
