Fig. 2.
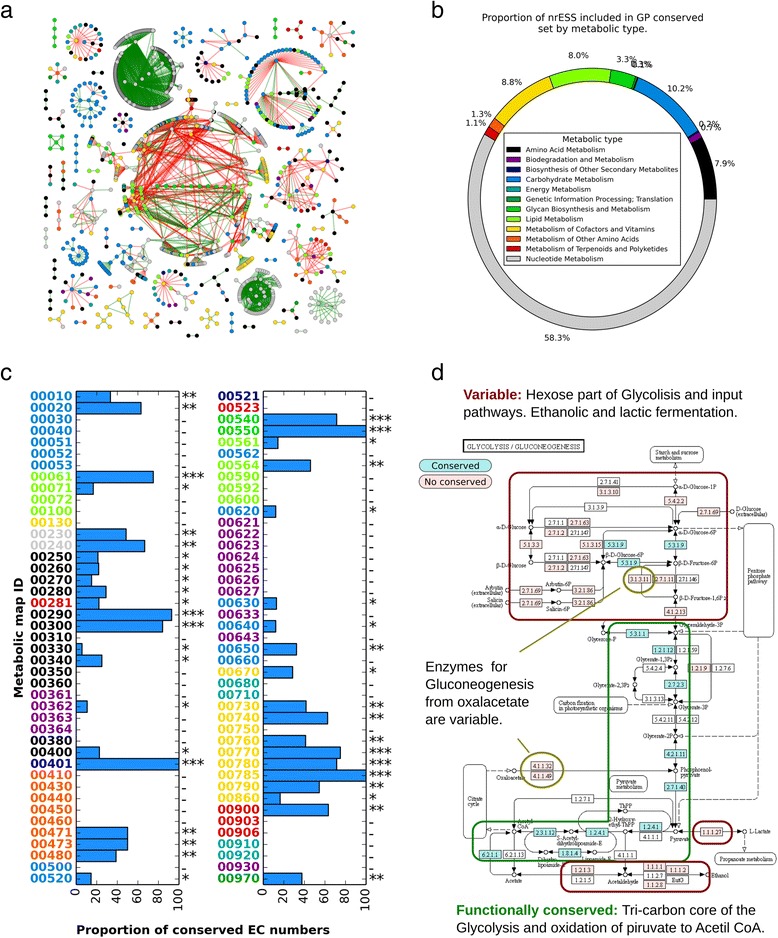
Functional conserved and variable nrESS in Gammaproteobacteria. a Graph representation of the relationships between the set of conserved ESS in Gammaproteobacteria. The nodes represent the nrESS and the edges show the alignments among them. Green edges are alignments between the same metabolic map, and the red ones represent alignments between different metabolic maps. The nodes are colored according to the metabolism type, as indicated. The alignments between ESS from the same map were selected as the Metabolic Map Functional Conserved Dataset (MMFCD). b Proportion of ESS from each metabolic type included in the MMFCD. c Proportion of EC numbers conserved in the alignments of the MMFCD for each metabolic map. The metabolic maps are represented by the KEGG map ID, and the colors indicate the metabolic type. The metabolic maps were classified according to the proportion of functional conserved EC numbers, as follows: conserved (***), more than 70 % of EC numbers conserved; moderately conserved (**), between 30 and 69 % of numbers conserved; barely conserved (*), between 1 and 30 % of numbers conserved; non conserved (-), 0 % of numbers conserved. d Functional conservation of the glycolysis/gluconeogenesis metabolic map in Gammaproteobacteria. The map represents the functional mapping of conserved nrESS (3 levels of EC classification) in the alignments of the MMFCD (cyan). In addition, the nrESS present in at least one of the species analyzed but not in the conserved set are shown (pink). The white steps represent enzymes not present in any of the species analyzed. In boxes and circles are represented the conserved and variable regions in the metabolic map. See text for details
