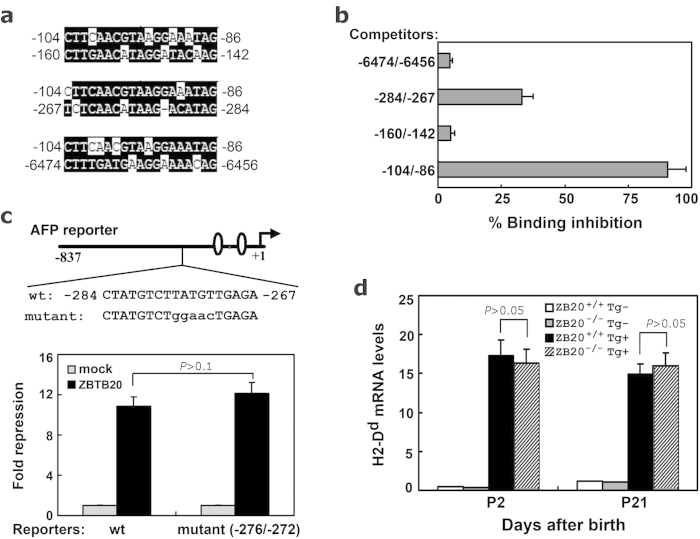
Figure 4. Analysis of other potential ZBTB20 sites in AFP gene.
(a) Sequence alignment of AFP gene fragment −104/−86 with −160/−142, −284/−267, and −6474/−6456, respectively. (b) Competitive ZBTB20-binding capacity of different AFP gene fragments in EDBA assay. 100-fold excess of AFP gene fragments competed with biotinylated fragment −104/−86 to bind to ZBTB20. (c) AFP −276/−272 mutant reporter was repressed by ZBTB20 as effectively as WT counterpart. Schematic demonstration of mutant AFP reporter was shown schematically in upper part, with the mutant nucleotides at −276/−272 of AFP gene reporter indicated in lower case. The reporter plasmids were cotransfected into HepG2 cells with mock control vector (gray bar) or ZBTB20-expressing plasmids (black bar). Results are expressed as fold repression of luciferase normalized to the internal control RL-SV40. n = 3 experiments. P > 0.1. (d) ZBTB20 ablation didn’t compromise the inhibitory activity of AFP Enhancer III in liver. Transgenic mice EIII-βgl-Dd were crossed to ZBTB20 global knockout mice, H2-Dd mRNA levels in liver were measured by real-time RT-PCR at the age of day 2 and day 21. ZBTB20+/+Tg− (open bar), ZBTB20−/−Tg− (gray bar), ZBTB20+/+Tg+ (black bar) ZBTB20−/−Tg+ (slashed bar). The transgenic expression did not differ significantly in ZBTB20-null livers. P > 0.05. n = 4 experiments.