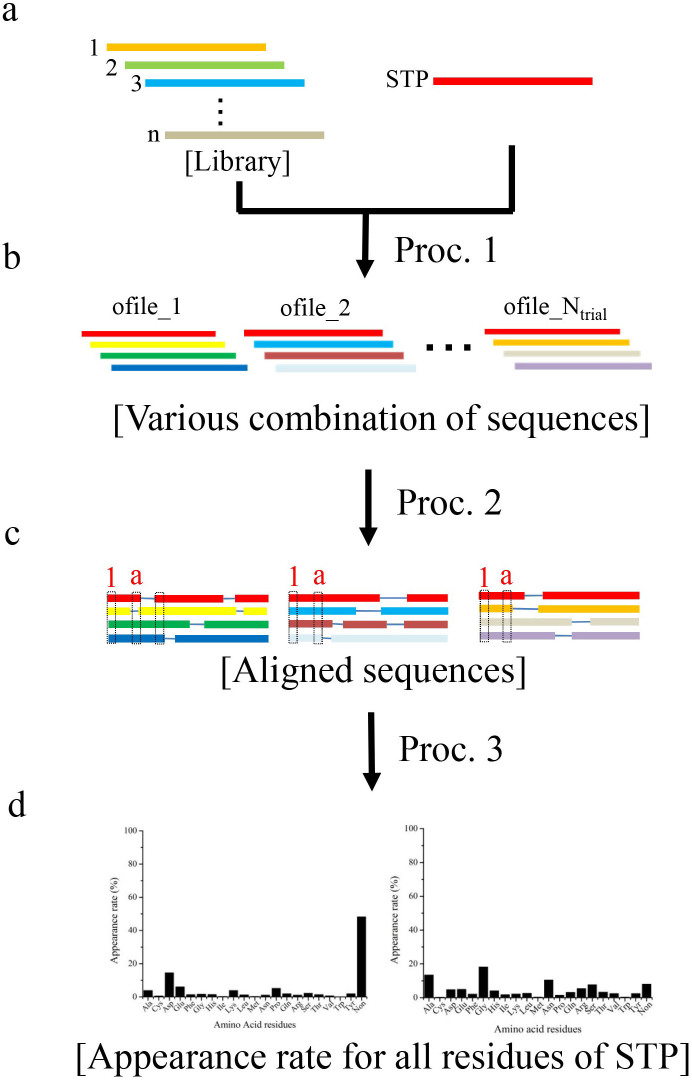
Figure 1. Schematic model of the INTMSAlign algorithm.
Sequence of target protein (STP) and library, which contains hundreds to thousands of sequences from the STP family, must be prepared before the INTMSAlign algorithm is used (a). After the preparation of the library, Ntrial ofiles are created by selecting one STP (red) and Npick sequences from the library (proc. 1). Here, the STP is contained in the ofile (b). Next, multiple sequence alignment is applied to all of the ofiles using ClustalW (proc. 2), and Ntrial of the aligned sequences are prepared (c). Finally, for Ntrial aligned sequences, the number of all 20 amino-acid residues and gaps is summed up (dotted square) for every STP residue (red color letter in c). Finally, the frequencies are calculated (d).