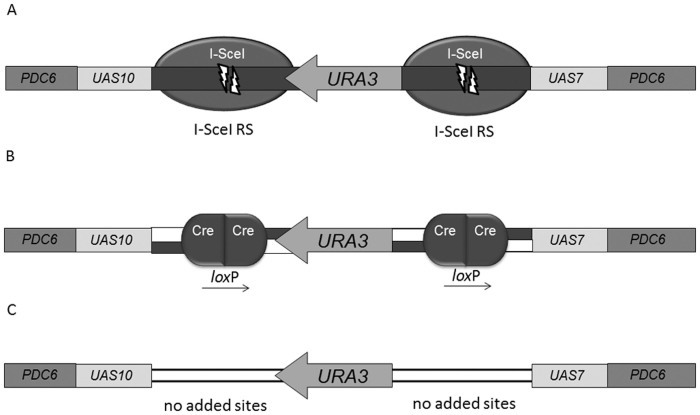
Figure 2. Target loci used to assay the effect of DSBs and nucleosome occupancy on the integration of T-DNA by HR.
These target loci were all integrated at the PDC6 locus, all containing sequence homology to the incoming T-strands by two different UAS type promoters derived from the yeast galactose pathway or by the flanking PDC6 fragments. To remove nucleosomes from these promoter sequences, the yeast cells are grown in a medium with galactose as the sole carbon source. Growth in a glucose medium would induce the formation of nucleosomes on the UAS typed promoters. The central URA3 marker is flanked by: (2A) recognition sites of the homing endonuclease I-SceI that catalyzes the formation of DSBs as a monomer, (2B) recognition sites for the recombinase Cre allowing for the removal of the URA3 marker gene without inducing double stranded break formation, (2C) target locus without any added recognition sites but still provided with homology to the incoming T-strands.