Figure 1. The post-transcriptional regulome of the 3′ UTR.
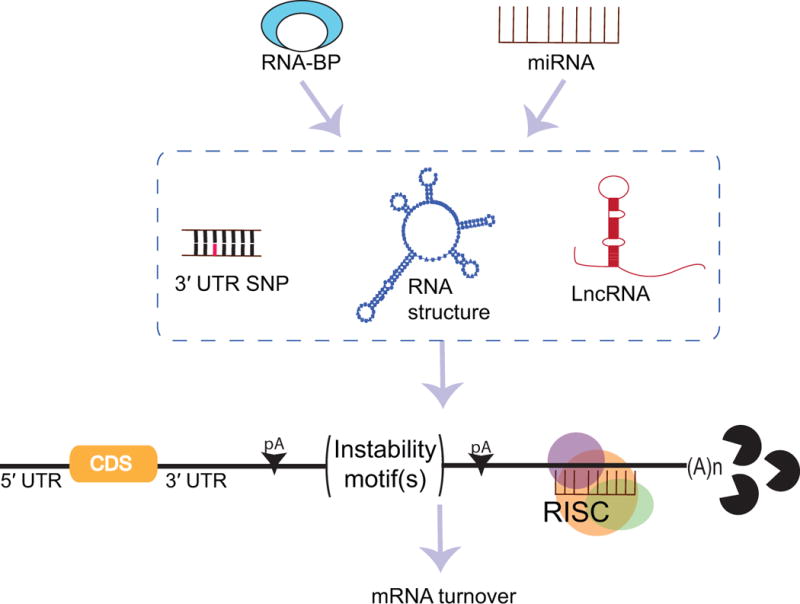
mRNA stability and gene expression is dictated by various post-transcriptional modulators interacting with the 3′-untranslated region (UTR). microRNAs (miRNAs) recruit the RNA-induced silencing complex (RISC) to specific target regions leading to ribonuclease-mediated mRNA decay. Similarly, instability motifs located within the 3′ UTR are targeted by RNA-binding proteins (RBPs), resulting in rapid poly(A) tail deadenylation and mRNA degradation, or in mRNA stabilization, respectively. Single nucleotide polymorphisms (SNPs) in the 3′ UTR disrupt the nucleotide complementarity needed for miRNA-mRNA interaction hence altering the binding capacity of miRNAs. In a similar fashion, SNPs can change the overall mRNA structure or particular instability motifs required for efficient RBP-mRNA interaction. Long non-coding RNAs (lncRNAs) are overarching modifiers of miRNA and RBP activity through sequestration, thereby suppressing their function. Finally, shortening of the 3′ UTR through usage of alternative polyadenylation sites (pA) affects overall mRNA stability by decreasing the number of potential interaction sites/motifs for the previously mentioned post-transcriptional modulators. These regulatory elements acting in sync define the post-transcriptional regulome of the 3′ UTR and ultimately dictate mRNA turnover and expression of a given gene.
