Fig. 1.
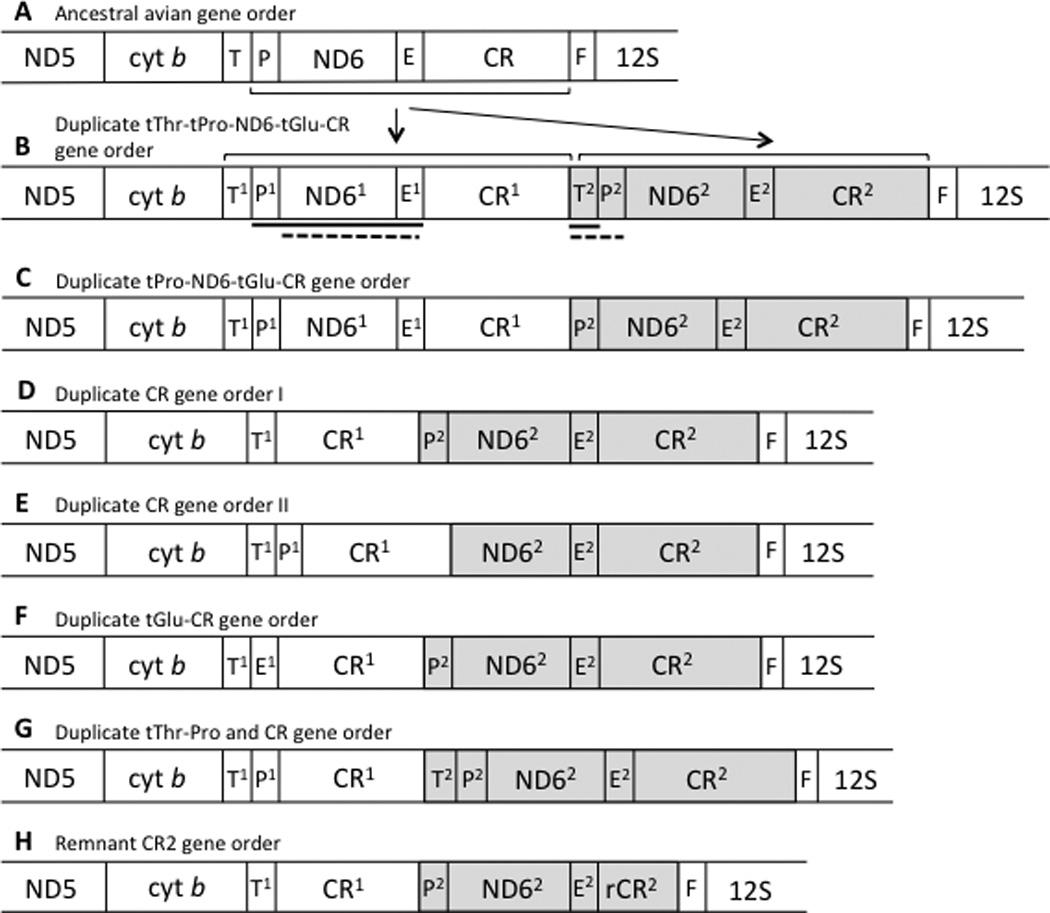
Gene orders surrounding the control region (CR) known for avian mitochondrial genomes. Brackets and arrows indicate the segment involved in a tandem duplication that has been hypothesized to be involved in the conversion between the standard and alternative gene orders. The solid and dashed lines beneath B indicate sections that degenerate (or are lost) in the transitions from B to D and E, respectively, to yield the two gene orders observed in parrots. Gene order H is similar to D, but the second copy of the control region is degenerate. Transfer RNAs are indicated by their single-letter abbreviations, E (Glutamyl), F (Phenylalanine), P (Proline), and T (Threonine). Pseudogenes, which have been described in various taxa for this portion of the mitochondrial genome, are not shown. Shading indicates hypothesized homology between highlighted regions. Taxa for which these different orders have been reported to date are listed in Table 1.
