FIG 4.
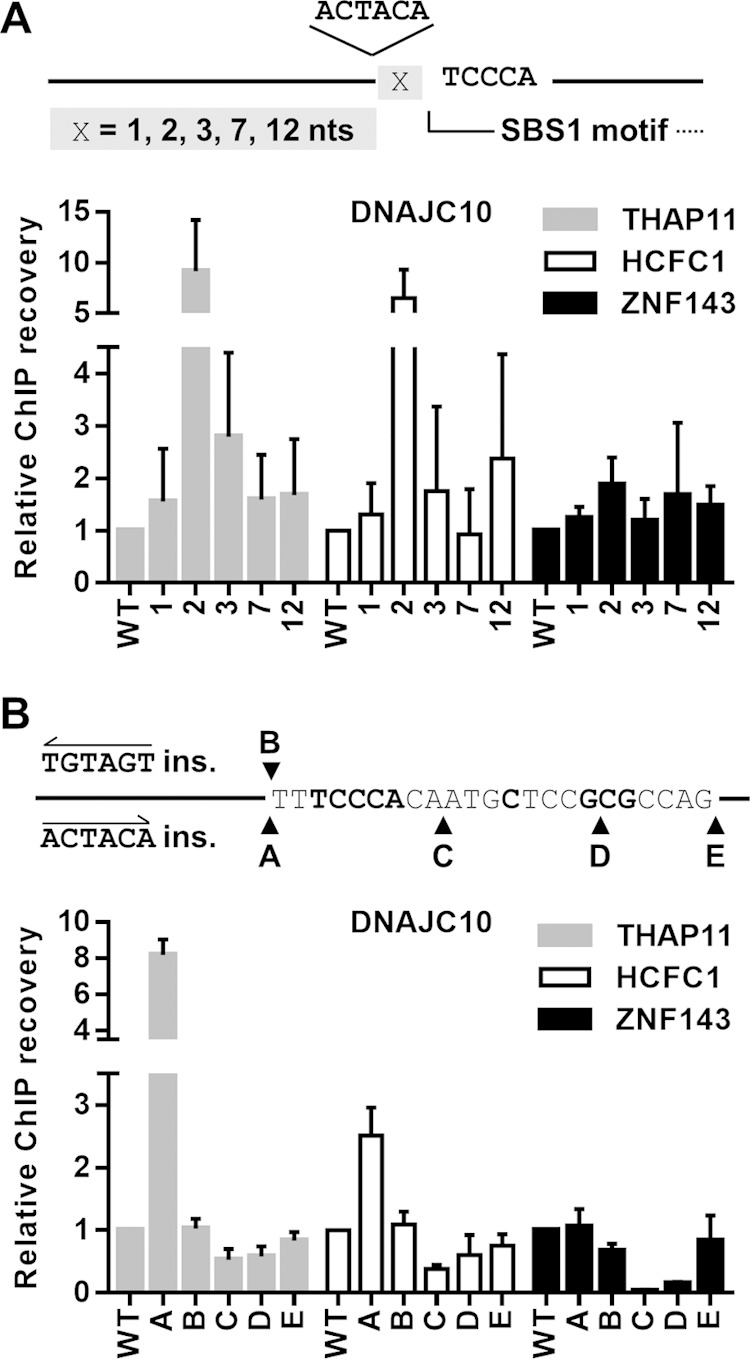
Two-nucleotide spacing, position, and orientation of the ACTACA sequence are critical for THAP11/HCFC1 recruitment. (A) Diagram depicting the location of the ACTACA sequence insertion relative to the SBS1 motif in the DNAJC10 promoter. X represents a single nucleotide. The ACTACA sequence was inserted 1, 2, 3, 7, or 12 nucleotides 5′ of the SBS1 motif. x-axis labels for bar graphs correspond to the numbers of nucleotides between the SBS1 motif and the ACTACA sequence. (B) Diagram depicting the locations and sequences that were inserted into the wild-type DNACJ10 retroviral construct. The labels shown for each insertion also correspond to the x-axis labels for bar graphs. The TGTAGT sequence inserted at the “B” position is the reverse complement of the ACTACA sequence and corresponds to the everted arrangement of the ACTACA sequence and SBS1 motif. Mutated and chromosomally integrated DNAJC10 promoters were assayed by ChIP using THAP11, HCFC1, and ZNF143 antibodies. Bar graphs indicate means ± standard deviations of data from 3 replicate ChIP experiments.
