Figure 1.
Loss of Cdx2 mRNA Selectively Impairs ISC Proliferation
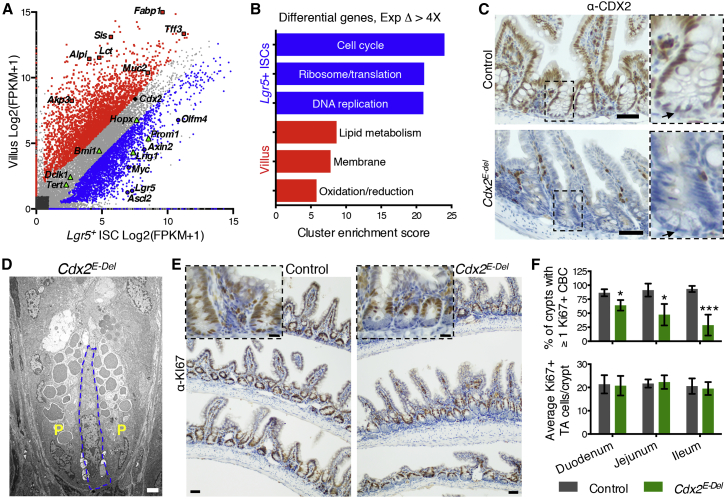
(A) RNA-seq data from three biological replicates of Lgr5+ ISCs and VCs identify modulated genes. Grey, 6,752 genes with comparable expression in both cell types; red, 3,317 VC-enriched genes; blue, 2,889 ISC-enriched genes (>2-fold; Q < 0.05). Markers of enterocytes or goblet cells (red), Lgr5+ ISCs (blue), and other ISC populations (green) are highlighted. Cdx2 is highly expressed in both cell types.
(B) Gene Ontology clusters of genes with >4-fold change in VCs and ISCs.
(C) CDX2 immunohistochemistry (IHC) in control and Cdx2E-Del mouse ileum 3 days after final TAM treatment, confirming complete epithelial loss, including ISCs (magnified to right; arrows).
(D) Electron microscopy of Cdx2E-Del ileum confirms the presence of ISCs (blue) among Paneth (P) cells.
(E) KI67 IHC reveals loss of proliferation in ISCs (magnified in Inset), but not in TA cells.
(F) Fraction of crypts with at least one KI67+ ISC in different regions of control and Cdx2E-Del intestines (∗p < 0.01; ∗∗∗p < 0.0001; t test). The average number of KI67+ TA cells is not significantly altered. At least 30–50 crypts were counted in each of at least four biological replicates.
Scale bars represent 50 μm (C and E), 20 μm (inset in E), and 2 μm (D).