Figure 3.
Genes Perturbed by CDX2 Loss and Delineation of CDX2 Binding in ISCs
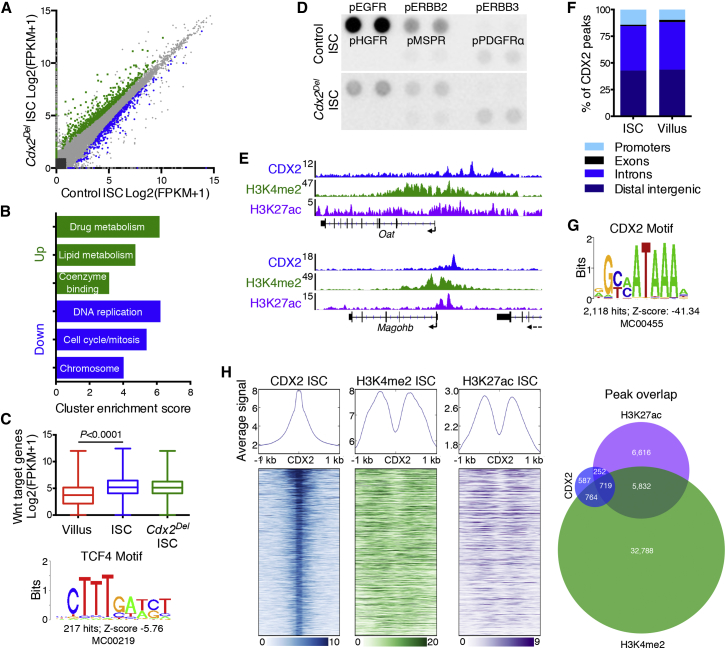
(A) RNA-seq data from isolated control and Cdx2Del ISCs, using three biological replicates. 639 transcripts are elevated in Cdx2−/− ISCs (green), and 412 transcripts are reduced (blue). Grey, genes with <1.5-fold change or Q > 0.05.
(B) Gene Ontology cluster enrichment among genes reduced or increased in Cdx2Del ISCs.
(C) Average RNA-seq FPKM values of 203 intestinal Wnt target genes in control VCs and ISCs and in Cdx2Del ISCs. Significance was assessed by a t test. Bottom: the TCF4 motif found in 217 CDX2 binding sites.
(D) Phospho-receptor tyrosine kinase arrays probed with lysates from purified control or Cdx2Del ISCs. Signals for pEGFR and pERBB2 are reduced in Cdx2Del ISCs.
(E) ChIP-seq signals for CDX2, H3K4me2, and H3K27ac near the Oat and Magohb genes; the y axis represents aligned ChIP-seq tag counts.
(F) Distribution of CDX2 binding sites among genomic regions is similar in ISCs and VCs.
(G) The consensus CDX2 motif is the most enriched (Z score: −41.34) in CDX2-bound sites.
(H) Heatmaps of ChIP signals for CDX2, H3K4me2, and H3K27ac around CDX2 binding sites in purified ISCs. Right: overlap of CDX2 binding sites with called H3K4me2 and H3K27ac peaks, which underestimate the overlap evident in heatmaps.