Abstract
Cefepime is frequently prescribed to treat infections caused by AmpC-producing Gram-negative bacteria. CMY-2 is the most common plasmid-mediated AmpC (pAmpC) β-lactamase. Unfortunately, CMY variants conferring enhanced cefepime resistance have been reported. Here, we describe the evolution of CMY-2 to an extended-spectrum AmpC (ESAC) in clonally identical Escherichia coli isolates obtained from a patient. The CMY-2-producing E. coli isolate (CMY-2-Ec) was isolated from a wound. Thirty days later, one CMY-33-producing E. coli isolate (CMY-33-Ec) was detected in a bronchoalveolar lavage fluid sample. Two weeks before the isolation of CMY-33-Ec, the patient received cefepime. CMY-33-Ec and CMY-2-Ec were identical by repetitive extragenic palindromic-PCR (rep-PCR), being of hyperepidemic sequence type 131 (ST131) but showing different β-lactam MICs (e.g., cefepime MIC, 16 and ≤0.5 μg/ml for CMY-33-Ec and CMY-2-Ec, respectively). Identical CMY-2-Ec isolates were also found in a rectal swab. CMY-33 differs from CMY-2 by a Leu293-Ala294 deletion. Expressed in E. coli strain DH10B, both CMYs conferred resistance to ceftazidime (≥256 μg/ml), but the cefepime MICs were higher for CMY-33 than CMY-2 (8 versus 0.25 μg/ml, respectively). The kcat/Km or inhibitor complex inactivation (kinact)/Ki app (μM−1 s−1) indicated that CMY-33 possesses an extended-spectrum β-lactamase (ESBL)-like spectrum compared to that of CMY-2 (e.g., cefoxitin, 0.2 versus 0.4; ceftazidime, 0.2 versus not measurable; cefepime, 0.2 versus not measurable; and tazobactam, 0.0018 versus 0.0009, respectively). Using molecular modeling, we show that a widened active site (∼4-Å shift) may play a significant role in enhancing cefepime hydrolysis. This is the first in vivo demonstration of a pAmpC that under cephalosporin treatment expands its substrate spectrum, resembling an ESBL. The prevalence of CMY-2-Ec isolates is rapidly increasing worldwide; therefore, awareness that cefepime treatment may select for resistant isolates is critical.
INTRODUCTION
Enterobacteriaceae can manifest resistance to third-generation cephalosporins as a result of the production of extended-spectrum β-lactamases (ESBLs), chromosomal AmpC (cAmpCs), or plasmid-mediated AmpCs (pAmpCs) (1, 2). In general, ESBLs are inhibited by the commercially available β-lactamase inhibitors but hydrolyze well the fourth-generation cephalosporin cefepime (FEP). On the other hand, AmpCs are not inhibited by inhibitors and do not hydrolyze FEP (1, 3–5). Therefore, FEP is suggested for the treatment of infections caused by AmpC producers (6–8).
In the past, AmpC variants with enhanced hydrolytic efficiency against FEP were sporadically reported in Enterobacter spp. (8–11), Serratia marcescens (12), and Escherichia coli (13–17). These chromosomal extended-spectrum AmpC β-lactamases (cESACs) possess specific amino acid insertions, deletions, duplications, or substitutions in the H-10 helix (also named the R2-loop) that allow better accommodation and hydrolysis of FEP in the serine active site (1, 4, 11, 14). More recently, plasmid-mediated ESACs (pESACs) derived from the most frequently detected pAmpC (CMY-2) were also identified (1). In particular, we previously reported the phenotypic characteristics of CMY-33- and CMY-44-producing E. coli isolates (18); further pESACs were described in E. coli (CMY-10 and CMY-94) and Klebsiella pneumoniae (CMY-19) (19–21). However, for these pESACs, structural information regarding the hydrolytic performance of different β-lactam substrates is still needed (20).
With regard to the possible factors leading to the generation of ESACs, treatment with β-lactams (especially third-generation cephalosporins and FEP) may permit the evolution of specific changes in the H-10 helix of the AmpC β-lactamase. However, in only three cases involving infections due to Enterobacter spp., the initial FEP-susceptible (FEPs) isolates were available for comparison with subsequent isolates expressing the cESAC variants after FEP treatment (8, 9, 11). To our knowledge, similar clinical cases have not yet been reported for strains producing pESAC enzymes. Here, we report such a case to illustrate the dynamic nature of this process.
MATERIALS AND METHODS
Clinical case.
A 71-year-old man underwent a radical cystectomy due to carcinoma of the urinary bladder. On day 14, an E. coli isolate (Ec-1) resistant to ceftriaxone (CRO) but susceptible to FEP was isolated in a swab taken from the surgical wound. One week later, the patient developed pneumonia that was empirically treated with CRO. After 2 days, the therapy was switched to FEP and continued for 6 days. Two weeks later, an E. coli isolate (Ec-2) resistant to both CRO and FEP was detected in a bronchoalveolar lavage fluid sample. A rectal swab also revealed that the patient was colonized with third-generation cephalosporin-resistant E. coli isolates; five colonies (named Ec-A to Ec-E) were randomly chosen from the selective plates for further investigations (see Text S1 in the supplemental material for a full description of the clinical case).
Phenotypic tests.
Species identification was achieved using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker). The rectal swab was enriched overnight in lysogenic broth (LB) containing cefuroxime (3 μg/ml) and then plated on BLSE, chromID ESBL (bioMérieux), and Supercarba selective agars (22); a disk of FEP was also placed at the center of these plates to detect FEP-resistant (FEPr) strains. MICs were obtained in cation-adjusted Mueller-Hinton II (MHII) broth (BBL) using microdilution ESB1F and GNX2F panels (Trek Diagnostics Systems) and interpreted according to the EUCAST criteria (23). The MICs for FEP were also measured using the Etest (bioMérieux) on MHII plates with (200 μg/ml) and without cloxacillin (Sigma) (1).
Characterization of resistance genes and clonality.
The CT103XL microarray (Check-Points) was used to screen for bla genes. PCR and DNA sequencing for acquired bla, ISEcp1, ISCR1, blacAmpC and its upstream region, and ompF and ompC porin genes were performed (18, 24–27). The results were compared to E. coli K-12 patterns (GenBank accession no. U00096). Genetic relatedness was studied using repetitive extragenic palindromic-PCR (rep-PCR) (28) and multilocus sequence typing (MLST) (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). Polymorphisms in the fimH and fimA genes of the type 1 fimbriae were also analyzed (29, 30).
Plasmid analysis.
Plasmid replicon typing was performed using the PBRT kit (Diatheva) (31). Plasmid extraction was achieved using the PureYield plasmid midiprep system (Promega). Conjugation was performed using NEB 5-α competent E. coli (New England BioLabs), and cells were selected on LB plates containing ampicillin (20 μg/ml) (Sigma) (32). The plasmid DNA of the transconjugants was restricted with EcoRV and PstI enzymes (Bio-Concept) (33).
Cloning of blaCMY genes, purification of proteins, and kinetic experiments.
blaCMY-2 and blaCMY-33 were cloned into pBC SK(−) and electroporated into E. coli strain DH10B cells (18). The MICs were obtained with the agar dilution method (34). The purification of CMY-33 was performed as previously done for CMY-2 (25, 35).
Steady-state kinetic analyses were performed on an Agilent 8453 spectrophotometer; the maximum rate of metabolism (Vmax) and Km for nitrocefin (NCF), cephalothin, and cefoxitin (FOX) were obtained using Origin 7.5 (OriginLab) (25). For the “poor substrates” (kcat, <2 s−1 or not measurable), cefotaxime (CTX), ceftazidime (CAZ), FEP, and aztreonam (ATM), the apparent Km was obtained as a competitive inhibition constant (Ki app) in the presence of NCF (36). For sulbactam (SUL) and tazobactam (TAZ), the inhibitor complex inactivation (kinact) in the presence of NCF was measured and the Ki app determined (25).
Molecular modeling and docking of cefepime.
The CMY-33 model was generated by the SWISS-MODEL server (http://swissmodel.expasy.org) using the deposited CMY-2 (PDB code 1zc2). The CMY-33 model was optimized by energy minimization (Discovery Studio 3.1 software; Accelrys) and using steepest descent and conjugate gradient algorithms to reach the minimum convergence (0.02 kcal/mol · Å). The protein was immersed in a water box (7 Å from any face), and the solvation model was used with periodic boundary conditions. The force field parameters of CHARMM were used for minimization, and the particle mesh Ewald method addressed long-range electrostatics. The bonds that involved hydrogen atoms were constrained with the SHAKE algorithm. CMY-33 model and CMY-2 were used for constructing the acylation complexes of both β-lactamases with FEP (25).
RESULTS AND DISCUSSION
In this case, the two E. coli isolates were resistant to third-generation cephalosporins but susceptible to piperacillin-tazobactam, carbapenems, and non-β-lactam antibiotics. However, while Ec-1 was susceptible to FEP (MIC, ≤0.5 μg/ml), Ec-2 was resistant to the drug, and its MIC was significantly reduced (from 16 to 1 μg/ml) in the presence of cloxacillin (Table 1). Since this behavior was suggestive of ESAC production (1), we characterized both E. coli isolates.
TABLE 1.
Phenotypic characterization of clinical isolates, transconjugants, and transformants producing the CMY-2 or CMY-33 plasmid-mediated AmpC β-lactamases
| Antibiotic | MIC (μg/ml) for E. colia: |
||||||
|---|---|---|---|---|---|---|---|
| Ec-1 isolate from wound (blaCMY-2) | Ec-2 isolate from BAL fluid (blaCMY-33) | NEB5α with IncI1 plasmid of Ec-1 (blaCMY-2) | NEB5α with IncI1 plasmid of Ec-2 (blaCMY-33) | DH10B pBC SK(−)/blaCMY-2b | DH10B pBC SK(−)/blaCMY-33b | DH10Bb | |
| Ampicillin | ≥32, R | ≥32, R | ≥32, R | ≥32, R | ≥512 | 128 | 4 |
| Ampicillin-sulbactam | NT | NT | NT | NT | 32 | 0.125 | 4 |
| Piperacillin | NT | NT | NT | NT | ≥512 | 32 | 2 |
| Piperacillin-tazobactam | ≤2, S | 8, S | ≤2, S | 16, S | 4 | 8 | 2 |
| Ticarcillin-clavulanate | ≤8, S | ≥256, R | ≤8, S | ≥256, R | NT | NT | NT |
| Cephalothin | ≥32, NA | ≥32, NA | ≥32, NA | ≥32, NA | ≥512 | 128 | 4 |
| Cefoxitin | 64, NA | 32, NA | 64, NA | ≥128, NA | 64 | 16 | 4 |
| Ceftriaxone | 8, R | 128, R | 16, R | ≥256, R | NT | NT | NT |
| Cefotaxime | 4, R | 64, R | 16, R | 64, R | 8 | 16 | ≤0.06 |
| Cefotaxime-clavulanate | 4, NA | 32, NA | 8, NA | 32, NA | NT | NT | |
| Ceftazidime | 8, R | ≥256, R | 32, R | ≥256, R | 256 | ≥512 | ≤0.06 |
| Ceftazidime-clavulanate | 4, NA | ≥256, NA | 16, NA | ≥256, NA | NT | NT | NT |
| Cefpodoxime | ≥64, R | ≥64, R | ≥64, R | ≥64, R | NT | NT | NT |
| Cefepime | ≤0.5, S (0.125/≤0.016) | 16, R (16/1) | ≤0.5, S | ≥32, R | 0.25 | 8 | ≤0.06 |
| Aztreonam | 4, I | ≥32, R | 8, I | ≥32, R | 8 | 16 | ≤0.06 |
| Imipenem | ≤0.25, S | ≤0.25, S | ≤0.25, S | ≤0.25, S | ≤0.5 | ≤0.5 | ≤0.5 |
| Meropenem | ≤0.5, S | ≤0.5, S | ≤0.5, S | ≤0.5, S | ≤0.06 | ≤0.06 | ≤0.06 |
| Ertapenem | ≤0.125, S | ≤0.125, S | ≤0.125, S | ≤0.125, S | NT | NT | NT |
| Gentamicin | ≤0.5, S | ≤0.5, S | ≤0.5, S | ≤0.5, S | NT | NT | NT |
| Tobramycin | ≤0.5, S | ≤0.5, S | ≤0.5, S | ≤0.5, S | NT | NT | NT |
| Amikacin | ≤2, S | ≤2, S | ≤2, S | ≤2, S | NT | NT | NT |
| Ciprofloxacin | ≤0.125, S | ≤0.125, S | ≤0.125, S | ≤0.125, S | NT | NT | NT |
| Doxycycline | ≤1, NA | ≤1, NA | ≤1, NA | ≤1, NA | NT | NT | NT |
| Tigecycline | ≤0.125, S | ≤0.125, S | ≤0.125, S | ≤0.125, S | NT | NT | NT |
| Co-trimoxazole | ≤0.25, S | ≤0.25, S | ≤0.25, S | ≤0.25, S | NT | NT | NT |
| Colistin | ≤0.125, S | ≤0.125, S | ≤0.125, S | ≤0.125, S | NT | NT | NT |
BAL, bronchoalveolar lavage; R, resistant; NT, not tested; S, susceptible; NA, not available; I, intermediate. The MICs were obtained with microdilution Trek panels and interpreted according to the EUCAST criteria (23). The tests were repeated three times. Values in parentheses indicate MICs obtained with the Etest method on plates without/with cloxacillin (200 μg/ml).
MICs for E. coli DH10B transformants containing pBC SK(−) were achieved with the agar dilution method. Consistent results were also previously obtained with the Etest method (18).
Molecular characterization.
Ec-1 possessed the blaCMY-2 gene (CMY-2-Ec), whereas Ec-2 harbored the blaCMY-33 gene (CMY-33-Ec); both blaCMY genes were located downstream of a truncated ISEcp1 (ΔISEcp1-3′), an element commonly associated with this group of genes (24, 37, 38). Other acquired bla genes were not detected. Ec-1 and Ec-2 also possessed (i) an identical promoter region of the blacAmpC with specific mutations (+81A, −28A, −73T, and −118A) leading to overproduction of the enzymes (15, 24); (ii) a cAmpC protein with Ala8Thr, Lys40Arg, Gln191Lys, Pro209Ser, Thr263Ile, Ser298Ile, Ala316Pro, Asp367Thr, and Ala375Thr; (iii) OmpF with several substitutions (identical to GenBank accession no. HG941718); and (iv) disrupted OmpC (identical to GenBank accession no. HG941718).
Both Ec-1 and Ec-2 carried plasmid replicon types I1, FIB, and FII. Each NEB 5-α transconjugant possessing the corresponding blaCMY (along with the replicon type IncI1) displayed phenotypic patterns consistent with the specific CMY produced (Table 1). Moreover, plasmid extracts from these transconjugants generated identical restriction patterns (see Fig. S1 in the supplemental material). Thus, we concluded that both blaCMY variants were carried in the same IncI1 plasmid.
Remarkably, all five strains obtained from the rectal swab (Ec-A to Ec-E) were CMY-2-Ec, whereas CMY-33-Ec was not detected. As 80% of FEP is excreted intact in urine (3), selective pressure upon the intestinal flora may be limited, which might explain why we did not find CMY-33-Ec strains in the rectal specimen, even after the administration of FEP.
The prevalence of CMY-2-Ec is rapidly increasing worldwide in human, food animal, and food chain settings, as the blaCMY genes are usually carried by epidemic conjugative plasmids (24, 38–41). In Switzerland, the rate of clinical specimens with CMY-2-Ec among those resistant to third-generation cephalosporins was 12.5% in 2011 (24). CMY-2-Ec pathogens can also be responsible for intestinal colonization in healthy people (42). Therefore, since Switzerland is among the major consumers of FEP in Europe, this situation may create in the near future the “perfect storm” to select more isolates producing pESAC variants of CMY-2 (43).
Genetic relatedness of E. coli isolates.
Ec-1, Ec-2, and the five CMY-2-Ec strains from the rectal swab had identical rep-PCR profiles (see Fig. S2 in the supplemental material). Moreover, all strains were of ST131 and possessed the fimH22/fimA7 type (30). Therefore, under selective pressure with CRO and mainly FEP, CMY-33 evolved from CMY-2 in the same E. coli host (the strain was colonizing the intestinal tract of the patient). E. coli ST131 is a hyperepidemic clone that has driven the worldwide spread of clinically important ESBLs (e.g., CTX-M-15) in both hospital and community settings (37). Consequently, the finding of a ST131 CMY-2-Ec (FEPs) that can evolve to those producing CMY-33 (FEPr) is a matter of serious clinical concern.
Biochemical characterization of CMY-2 and CMY-33.
Both CMYs expressed in E. coli DH10B cells conferred resistance to third-generation cephalosporins. However, cells producing CMY-33 had a phenotype resembling that of an ESBL producer because they had higher MICs for FEP but lower MICs for FOX and ampicillin-SUL than those producing CMY-2 (Table 1).
CMY-33 (GenBank accession no. EU496816) differs from CMY-2 (GenBank accession no. X91840) by a Leu293-Ala294 deletion in the H-10 helix. Electrospray ionization-mass spectrometry (ESI-MS) analysis indicated that the molecular mass of CMY-33 (39,671 Da) is less than that of CMY-2 (39,854 Da) due to the Leu293-Ala294 deletion (see Fig. S3 in the supplemental material). Consistent with the MIC determinations for the E. coli DH10B transformants, the steady-state kinetic parameters revealed that CMY-33 possesses significantly less catalytic efficiency (kcat/Km) for NCF and narrow-spectrum cephalosporins (cephalothin and FOX), but it is more inhibited (due to lower Ki app values of approximately 30 to 35%) by SUL and TAZ than by CMY-2. On the other hand, CMY-33 had improved hydrolytic activity against ATM, CTX, CAZ, and FEP, substrates for which detectable hydrolysis was not recorded for CMY-2 (Table 2) (25). Overall, these kinetic parameters support again that CMY-33 is a pESAC with a phenotype very reminiscent of classic ESBLs (5).
TABLE 2.
Steady-state kinetic parameters of purified CMY-2 and CMY-33 plasmid-mediated AmpC β-lactamasesa
| β-Lactamb |
Km or Ki
app (μM)c |
kcat or kinact for inhibitors (s−1) |
kcat /Km or kinact/Ki
app for inhibitors (μM−1 s−1) |
|||
|---|---|---|---|---|---|---|
| CMY-2 | CMY-33 | CMY-2 | CMY-33 | CMY-2 | CMY-33 | |
| NCF | 11.2 | 3.4 | 534.8 | 3.1 | 47.6 | 0.9 |
| CEF | 7.8 | 1.7 | 140.0 | 0.8 | 17.9 | 0.5 |
| FOX | 17.9 | 22.9 | 6.8 | 4.7 | 0.4 | 0.2 |
| CAZc | NMd | 20.0 | NM | 3.2 | NM | 0.2 |
| CTXc | 1.8 | 3.9 | NM | 3.2 | NM | 0.8 |
| FEPc | 108.1 | 18.3 | NM | 3.0 | NM | 0.2 |
| ATMc | 0.12 | 1.5 | NM | 4.2 | NM | 2.8 |
| SUL | 101.3 | 35.0 | 0.025 | 0.025 | 0.0002 | 0.0007 |
| TAZ | 50.0 | 16.6 | 0.045 | 0.028 | 0.0009 | 0.0018 |
Experimental error was ±10%.
NCF, nitrocefin; CEF, cephalothin; FOX, cefoxitin; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; SUL, sulbactam; TAZ, tazobactam.
Ki app measured in the presence of NCF. The data regarding CMY-2 are from Endimiani et al. (25).
NM, not measurable.
Molecular modeling and acylation complexes with FEP.
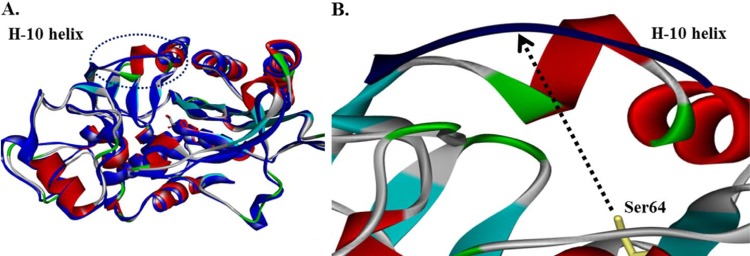
In our model, the deletion in the H-10 helix increases the distance from Ser64 to the H-10 helix by about 4 Å, altering the conformation, size, and possibly the flexibility of the active site of CMY-33 (Fig. 1). This may account for the differences in kinetic parameters. Previous hypotheses were raised stating that disruptions in the H-10 helix of cAmpCs (involving positions 282 to 296) are responsible for the ESBL phenotype. In particular, increased resistance to FEP, CAZ, CTX, and ATM was constantly observed in all previous cESACs (4, 11, 14); however, this is the first time that this phenomenon has been documented for a pESAC.
FIG 1.
(A) CMY-2 and CMY-33 model (blue) molecular structure superimposi tion. (B) Magnification of the H-10 helix in CMY-2 and CMY-33. The deletion (Leu293-Ala294) in the CMY-33 enzyme increases the distance from Ser64 to the H-10 helix by ~4 Å (from 7.8 to 11.6 Å), changing the shape, size, and possibly the flexibility of the active site.
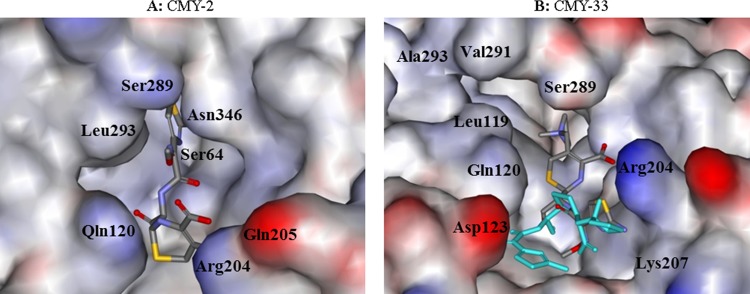
Our model suggests that the reason for increased hydrolysis of these substrates in CMY-33 is the ready formation of a Michaelis complex (kcat is greater, Km is lower). In particular, docking of FEP indicated that at least two different conformations are possible in the widened active site of CMY-33 rather than only one in CMY-2 (Fig. 2).
FIG 2.
Molecular representation of the acyl-enzyme complex of CMY-2 (A) and CMY-33 (B) as Connolly surface, with FEP docked in the active site. The model suggests that the reason for increased drug hydrolysis is the ready formation of Michaelis complex (kcat is greater, Km is lower). In particular, two different possible conformations were seen in the widened active site of CMY-33, suggesting the structural basis for resistance in CMY-33 versus CMY-2.
Conclusions.
We describe here the first clinical case in which a pAmpC (CMY-2) evolved to a pESAC (CMY-33) under FEP treatment. In particular, a ST131 FEPs CMY-2-Ec isolate rapidly became FEPr due to a double amino acid deletion in the H-10 helix of the protein. CMY-33 is an atypical pAmpC that mimics an ESBL (i.e., is relatively susceptible to standard inhibitors and FOX but resistant to oxyiminocephalosporins) and therefore classifiable as pESAC. Given that the prevalence of CMY-2-Ec isolates is rapidly increasing worldwide, one should be aware that the standard FEP treatment may select for resistant isolates in vivo.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Swiss National Science Foundation (SNF) (grant 32003B_153377 to A.E.). J.P. is a Ph.D. student (2014 to 2017) supported by the SNF. The research reported in this publication was also supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (grants R01AI063517 and R01AI100560 to R.A.B., and grants R21AI107302 and R01AI104895 to Y.D.); the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program (award 1I01BX001974 to R.A.B.); and the Geriatric Research Education and Clinical Center VISN 10 (to R.A.B.).
The content of this article is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
We thank Sara Droz (Institute for Infectious Diseases, University of Bern) for providing the E. coli isolates (Ec-1 and Ec-2) and Thierry Parret and Gwendoline Boillat (Hôpital du Jura Bernois SA, St-Imier, Switzerland) for the clinical data regarding the patient.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01804-15.
REFERENCES
- 1.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez F, Endimiani A, Hujer KM, Bonomo RA. 2007. The continuing challenge of ESBLs. Curr Opin Pharmacol 7:459–469. doi: 10.1016/j.coph.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endimiani A, Perez F, Bonomo RA. 2008. Cefepime: a reappraisal in an era of increasing antimicrobial resistance. Expert Rev Anti Infect Ther 6:805–824. doi: 10.1586/14787210.6.6.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vakulenko SB, Golemi D, Geryk B, Suvorov M, Knox JR, Mobashery S, Lerner SA. 2002. Mutational replacement of Leu-293 in the class C Enterobacter cloacae P99 β-lactamase confers increased MIC of cefepime. Antimicrob Agents Chemother 46:1966–1970. doi: 10.1128/AAC.46.6.1966-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Gniadkowski M, Nordmann P. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J Antimicrob Chemother 50:1031–1034. doi: 10.1093/jac/dkf240. [DOI] [PubMed] [Google Scholar]
- 6.Harris PN, Ferguson JK. 2012. Antibiotic therapy for inducible AmpC β-lactamase-producing Gram-negative bacilli: what are the alternatives to carbapenems, quinolones and aminoglycosides? Int J Antimicrob Agents 40:297–305. doi: 10.1016/j.ijantimicag.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Siedner MJ, Galar A, Guzmán-Suarez BB, Kubiak DW, Baghdady N, Ferraro MJ, Hooper DC, O'Brien TF, Marty FM. 2014. Cefepime vs other antibacterial agents for the treatment of Enterobacter species bacteremia. Clin Infect Dis 58:1554–1563. doi: 10.1093/cid/ciu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilty M, Sendi P, Seiffert SN, Droz S, Perreten V, Hujer AM, Bonomo RA, Muhlemann K, Endimiani A. 2013. Characterisation and clinical features of Enterobacter cloacae bloodstream infections occurring at a tertiary care university hospital in Switzerland: is cefepime adequate therapy? Int J Antimicrob Agents 41:236–249. doi: 10.1016/j.ijantimicag.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnaud G, Benzerara Y, Gravisse J, Raskine L, Sanson-Le Pors MJ, Labia R, Arlet G. 2004. Selection during cefepime treatment of a new cephalosporinase variant with extended-spectrum resistance to cefepime in an Enterobacter aerogenes clinical isolate. Antimicrob Agents Chemother 48:1040–1042. doi: 10.1128/AAC.48.3.1040-1042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnaud G, Labia R, Raskine L, Sanson-Le Pors MJ, Philippon A, Arlet G. 2001. Extension of resistance to cefepime and cefpirome associated to a six amino acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol Lett 195:185–190. doi: 10.1111/j.1574-6968.2001.tb10519.x. [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez-Martínez JM, Fernández-Echauri P, Fernandez-Cuenca F, Diaz de Alba P, Briales A, Pascual A. 2012. Genetic characterization of an extended-spectrum AmpC cephalosporinase with hydrolysing activity against fourth-generation cephalosporins in a clinical isolate of Enterobacter aerogenes selected in vivo. J Antimicrob Chemother 67:64–68. doi: 10.1093/jac/dkr423. [DOI] [PubMed] [Google Scholar]
- 12.Mammeri H, Poirel L, Bemer P, Drugeon H, Nordmann P. 2004. Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC β-lactamase of a Serratia marcescens clinical isolate. Antimicrob Agents Chemother 48:716–720. doi: 10.1128/AAC.48.3.716-720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mammeri H, Poirel L, Nordmann P. 2007. Extension of the hydrolysis spectrum of AmpC β-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. J Antimicrob Chemother 60:490–494. doi: 10.1093/jac/dkm227. [DOI] [PubMed] [Google Scholar]
- 14.Doi Y, Wachino J, Ishiguro M, Kurokawa H, Yamane K, Shibata N, Shibayama K, Yokoyama K, Kato H, Yagi T, Arakawa Y. 2004. Inhibitor-sensitive AmpC β-lactamase variant produced by an Escherichia coli clinical isolate resistant to oxyiminocephalosporins and cephamycins. Antimicrob Agents Chemother 48:2652–2658. doi: 10.1128/AAC.48.7.2652-2658.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haenni M, Châtre P, Madec JY. 2014. Emergence of Escherichia coli producing extended-spectrum AmpC β-lactamases (ESAC) in animals. Front Microbiol 5:53. doi: 10.3389/fmicb.2014.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillon H, Tande D, Mammeri H. 2011. Emergence of ertapenem resistance in an Escherichia coli clinical isolate producing extended-spectrum β-lactamase AmpC. Antimicrob Agents Chemother 55:4443–4446. doi: 10.1128/AAC.01513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crémet L, Caroff N, Giraudeau C, Dauvergne S, Lepelletier D, Reynaud A, Corvec S. 2010. Occurrence of ST23 complex phylogroup A Escherichia coli isolates producing extended-spectrum AmpC β-lactamase in a French hospital. Antimicrob Agents Chemother 54:2216–2218. doi: 10.1128/AAC.01580-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi Y, Paterson DL, Adams-Haduch JM, Sidjabat HE, O'Keefe A, Endimiani A, Bonomo RA. 2009. Reduced susceptibility to cefepime among Escherichia coli clinical isolates producing novel variants of CMY-2 β-lactamase. Antimicrob Agents Chemother 53:3159–3161. doi: 10.1128/AAC.00133-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wachino J, Kurokawa H, Suzuki S, Yamane K, Shibata N, Kimura K, Ike Y, Arakawa Y. 2006. Horizontal transfer of blaCMY-bearing plasmids among clinical Escherichia coli and Klebsiella pneumoniae isolates and emergence of cefepime-hydrolyzing CMY-19. Antimicrob Agents Chemother 50:534–541. doi: 10.1128/AAC.50.2.534-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JY, Jung HI, An YJ, Lee JH, Kim SJ, Jeong SH, Lee KJ, Suh PG, Lee HS, Lee SH, Cha SS. 2006. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase. Mol Microbiol 60:907–916. doi: 10.1111/j.1365-2958.2006.05146.x. [DOI] [PubMed] [Google Scholar]
- 21.Crémet L, Caroff N, Giraudeau C, Reynaud A, Caillon J, Corvec S. 2013. Detection of clonally related Escherichia coli isolates producing different CMY β-lactamases from a cystic fibrosis patient. J Antimicrob Chemother 68:1032–1035. doi: 10.1093/jac/dks520. [DOI] [PubMed] [Google Scholar]
- 22.Nordmann P, Girlich D, Poirel L. 2012. Detection of carbapenemase producers in Enterobacteriaceae by use of a novel screening medium. J Clin Microbiol 50:2761–2766. doi: 10.1128/JCM.06477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2015. Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf. [Google Scholar]
- 24.Seiffert SN, Hilty M, Kronenberg A, Droz S, Perreten V, Endimiani A. 2013. Extended-spectrum cephalosporin-resistant Escherichia coli in community, specialized outpatient clinic and hospital settings in Switzerland. J Antimicrob Chemother 68:2249–2254. [DOI] [PubMed] [Google Scholar]
- 25.Endimiani A, Doi Y, Bethel CR, Taracila M, Adams-Haduch JM, O'Keefe A, Hujer AM, Paterson DL, Skalweit MJ, Page MG, Drawz SM, Bonomo RA. 2010. Enhancing resistance to cephalosporins in class C β-lactamases: impact of Gly214Glu in CMY-2. Biochemistry 49:1014–1023. doi: 10.1021/bi9015549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Andrea MM, Nucleo E, Luzzaro F, Giani T, Migliavacca R, Vailati F, Kroumova V, Pagani L, Rossolini GM. 2006. CMY-16, a novel acquired AmpC-type β-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy. Antimicrob Agents Chemother 50:618–624. doi: 10.1128/AAC.50.2.618-624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oteo J, Delgado-Iribarren A, Vega D, Bautista V, Rodriguez MC, Velasco M, Saavedra JM, Pérez-Vázquez M, García-Cobos S, Martínez-Martínez L, Campos J. 2008. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int J Antimicrob Agents 32:534–537. doi: 10.1016/j.ijantimicag.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Hilty M, Betsch BY, Bögli-Stuber K, Heiniger N, Stadler M, Küffer M, Kronenberg A, Rohrer C, Aebi S, Endimiani A, Droz S, Mühlemann K. 2012. Transmission dynamics of extended-spectrum β-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis 55:967–975. doi: 10.1093/cid/cis581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul S, Linardopoulou EV, Billig M, Tchesnokova V, Price LB, Johnson JR, Chattopadhyay S, Sokurenko EV. 2013. Role of homologous recombination in adaptive diversification of extraintestinal Escherichia coli. J Bacteriol 195:231–242. doi: 10.1128/JB.01524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Endimiani A, Luzzaro F, Perilli M, Lombardi G, Coli A, Tamborini A, Amicosante G, Toniolo A. 2004. Bacteremia due to Klebsiella pneumoniae isolates producing the TEM-52 extended-spectrum β-lactamase: treatment outcome of patients receiving imipenem or ciprofloxacin. Clin Infect Dis 38:243–251. doi: 10.1086/380645. [DOI] [PubMed] [Google Scholar]
- 33.Carattoli A, Seiffert SN, Schwendener S, Perreten V, Endimiani A. 2015. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. doi: 10.1371/journal.pone.0123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed CLSI document M7-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.Thomson JM, Distler AM, Prati F, Bonomo RA. 2006. Probing active site chemistry in SHV β-lactamase variants at Ambler position 244. Understanding unique properties of inhibitor resistance. J Biol Chem 281:26734–26744. [DOI] [PubMed] [Google Scholar]
- 36.Papp-Wallace KM, Mallo S, Bethel CR, Taracila MA, Hujer AM, Fernandez A, Gatta JA, Smith KM, Xu Y, Page MG, Desarbre E, Bou G, Bonomo RA. 2014. A kinetic analysis of the inhibition of FOX-4 β-lactamase, a plasmid-mediated AmpC cephalosporinase, by monocyclic β-lactams and carbapenems. J Antimicrob Chemother 69:682–690. doi: 10.1093/jac/dkt434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seiffert SN, Hilty M, Perreten V, Endimiani A. 2013. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug Resist Updat 16:22–45. doi: 10.1016/j.drup.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Seiffert SN, Tinguely R, Lupo A, Neuwirth C, Perreten V, Endimiani A. 2013. High prevalence of extended-spectrum-cephalosporin-resistant Enterobacteriaceae in poultry meat in Switzerland: emergence of CMY-2- and VEB-6-possessing Proteus mirabilis. Antimicrob Agents Chemother 57:6406–6408. doi: 10.1128/AAC.01773-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Endimiani A, Rossano A, Kunz D, Overesch G, Perreten V. 2012. First countrywide survey of third-generation cephalosporin-resistant Escherichia coli from broilers, swine, and cattle in Switzerland. Diagn Microbiol Infect Dis 73:31–38. doi: 10.1016/j.diagmicrobio.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos AP, Mevius D, Peixe L, Poirel L, Schuepbach-Regula G, Torneke K, Torren-Edo J, Torres C, Threlfall J. 2013. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis 56:1030–1037. doi: 10.1093/cid/cis1043. [DOI] [PubMed] [Google Scholar]
- 42.Garrido A, Seral C, Gude MJ, Casado C, Gonzalez-Dominguez M, Saenz Y, Castillo FJ. 2014. Characterization of plasmid-mediated β-lactamases in fecal colonizing patients in the hospital and community setting in Spain. Microb Drug Resist 20:301–304. doi: 10.1089/mdr.2013.0109. [DOI] [PubMed] [Google Scholar]
- 43.Plüss-Suard C, Pannatier A, Kronenberg A, Mühlemann K, Zanetti G. 2011. Hospital antibiotic consumption in Switzerland: comparison of a multicultural country with Europe. J Hosp Infect 79:166–171. doi: 10.1016/j.jhin.2011.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.