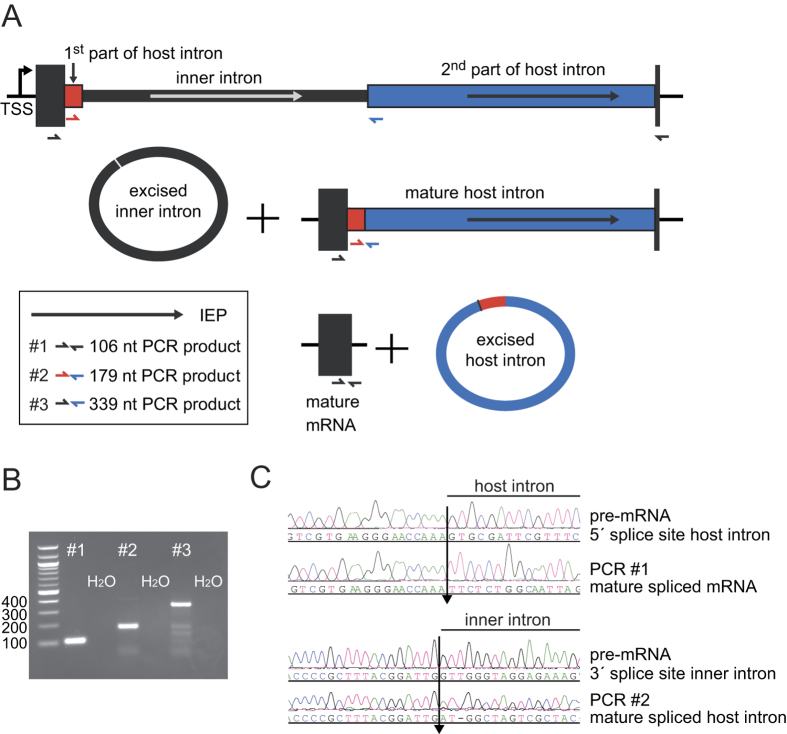
Figure 1. A twintron is present in the genome of T. erythraeum IMS101 and is actively spliced.
(A) Schematic representation of the twintron and its proposed splicing steps. In the scheme from left to right: arrow =TSS of the pre-mRNA, dark box = interrupted ORF Tery_4732, red box = 1st part of host intron, thin black box: inner intron interrupting domain 1 of the host intron, blue box = 2nd part of host intron, dark short dark box = 3′ exon of Tery_4732 mRNA. Arrows indicate the IEP ORFs Tery_4730 and Tery_4731 in the two introns (the IEPs share 30%/47% identical/similar residues with each other). The half-arrows aligned to the scheme indicate the primers used. The inset legend shows color-coded primer combinations used for the different amplicons. (B) #1 RT-PCR with primers in the 5′ (Tery_4732) and the 3′ mRNA-exon (black) yielded a product with the mature mRNA sequence (106 nt). #2 primers in the host intron (red and blue) yielded a product with the mature host intron sequence (179 nt, inner intron spliced out). #3 primers in Tery_4732 and the 2nd part of the host intron (black forward and blue reverse) verified the existence of an intermediate precursor without the inner intron (339 nt). H2O denotes control PCR without template cDNA. (C) Sequencing traces of amplicons #1 and #2, each compared to the respective unspliced sequence from the pre-mRNA. The arrows mark the identified splice sites. Note that the upper two sequences are in forward orientation, the lower two in reverse.