Abstract
Notch signaling is an evolutionary conserved process that influences cell fate determination, cell proliferation, and cell death in a context-dependent manner. Notch signaling is fine-tuned at multiple levels and misregulation of Notch has been implicated in a variety of human diseases. We have characterized maheshvara (mahe), a novel gene in Drosophila melanogaster that encodes a putative DEAD box protein that is highly conserved across taxa and belongs to the largest group of RNA helicase. A dynamic pattern of mahe expression along with the maternal accumulation of its transcripts is seen during early stages of embryogenesis. In addition, a strong expression is also seen in the developing nervous system. Ectopic expression of mahe in a wide range of tissues during development results in a variety of defects, many of which resemble a typical Notch loss-of-function phenotype. We illustrate that ectopic expression of mahe in the wing imaginal discs leads to loss of Notch targets, Cut and Wingless. Interestingly, Notch protein levels are also lowered, whereas no obvious change is seen in the levels of Notch transcripts. In addition, mahe overexpression can significantly rescue ectopic Notch-mediated proliferation of eye tissue. Further, we illustrate that mahe genetically interacts with Notch and its cytoplasmic regulator deltex in trans-heterozygous combination. Coexpression of Deltex and Mahe at the dorso-ventral boundary results in a wing-nicking phenotype and a more pronounced loss of Notch target Cut. Taken together we report identification of a novel evolutionary conserved RNA helicase mahe, which plays a vital role in regulation of Notch signaling.
Keywords: Drosophila, DEAD box helicase, Notch signaling, RNA-binding protein, RNA helicase
NOTCH signaling is an evolutionary conserved process that mediates cell–cell communication, which ultimately regulates cell fate (Artavanis-Tsakonas et al. 1999). Notch signaling is critical for many developmental processes and aberrant notch signaling has been related to many human diseases including cancer (Gridley 2003). Notch encodes a trans-membrane receptor that comprises an extracellular domain (NECD) and an intracellular domain (NICD). Notch is expressed at the cell surface as a heterodimeric receptor that is a result of furin-dependent cleavage (S1) occurring in the trans-Golgi network. At the cell surface it physically interacts with the ligands that are expressed in the apposing cells. This interaction with the ligand facilitates a series of proteolytic cleavages, ultimately resulting in the release of NICD. Released NICD translocates into the nucleus, where it interacts with a DNA binding protein CSL (mammalian CBF1/Drosophila Suppressor of Hairless/C. elegans Lag-1) and activates downstream gene expression, by relieving the repressor complex that silences Notch target genes (Artavanis-Tsakonas et al. 1983; Logeat et al. 1998; Struhl and Greenwald 1999; Brou et al. 2000; Kopan 2002; Lieber et al. 2002). Fine-tuning of Notch signaling is mediated by a vast number of Notch regulators or modifiers. Although Notch signaling has been extensively studied, identification of additional modifiers and regulators will help us to better understand the intricate regulation of this fundamental signaling pathway (Hall et al. 2004; Saj et al. 2010).
The RNA-binding protein of the DEAD box family constitutes the largest group of RNA helicases with signature motif DEAD (amino acid code for Asp, Glu, Ala, and Asp). They are known to unwind RNA in an ATP-dependent manner and can modulate RNA–RNA as well as RNA–protein interactions, which may influence the expression, localization, and stability of the target RNAs (Linder et al. 1989; Liu et al. 2008). Previously a number of DEAD box helicases were identified and their role as post-transcriptional gene regulators has been highlighted in different studies. A vast number of these helicases have been reported to participate in RNA metabolism (Hirling et al. 1989; Bond et al. 2001; Rocak and Linder 2004; Sengoku et al. 2004).
In this study we have characterized maheshvara (mahe), a novel gene in Drosophila that encodes a putative DEAD box RNA helicase and was isolated from a P-element screen for genes affecting life span in Drosophila (M. Mutsuddi, unpublished results).
Amino acid sequence comparison of Mahe protein indicates the presence of highly conserved motifs with similarity to DEAD box helicase human DDX5, yeast DBP2, and fly Rm62. DBP2 and Rm62 have been previously reported as yeast and Drosophila orthologs of DDX5 (Caretti et al. 2007) .
We report that mahe, which was originally designated as CG10777 in FlyBase, shows a dynamic pattern of expression. It is expressed maternally and exhibits strong neuronal expression during embryogenesis and larval development. Ectopic expression of mahe results in a wide range of phenotypes. Interestingly, ectopic expression of mahe in the salivary gland and eye-antennal disc leads to massive reduction in size of these tissues. This prompted us to name CG10777 as maheshvara (mahe), after a powerful Hindu deity, who is worshiped as the destroyer and restorer of the world.
Overexpression of mahe also leads to loss of bristles in the scutellum and notching of the wing margins, all of which resemble a Notch loss-of-function phenotype. Ectopic expression of mahe at the dorsal/ventral boundary (D/V) of wing discs resulted in downregulation of Notch targets Cut and Wingless as well as depletion of Notch protein but not its transcripts. In addition, maheshvara showed strong genetic interaction with both mutant alleles of both Notch and deltex (dx). Thus, our report indicates the involvement of an RNA-binding protein Mahe in regulation of Notch signaling.
Materials and Methods
Sequence comparison, phylogeny tree building, and model structure generation
A cartoon illustrating the conserved motifs in Mahe was generated using DOG 2.0 software. Sequence alignment was done using the Clustal omega program and a phylogenetic tree was built using the Clustal X program. ExPASy Swiss model 2.0 was used for homology-based generation of structure and chimera software was used to label the motifs at a specific position. For alignment and generating the phylogenetic tree, sequences used were human DDX5 (GenBank accession no. NP_004387.1), mouse DDX5 (GenBank accession no. NP_031866.2), zebra fish DDX5 (GenBank accession no. NP_001296454.1), worm DDX17 (GenBank accession no. NP_001041134.1), yeast DBP2 (GenBank accession no. NP_014287.3), fly Rm62 (GenBank accession no. NP_001246939.1), and fly Mahe (CG10777; GenBank accession no. NP_572424.1).
Drosophila stocks and genetic crosses
Fly stocks were maintained at 25° ± 1° on standard food unless otherwise stated. The following fly stocks were used in the present study: w1118 was used in control crosses; N54l9, a null mutant of Notch; dx152, a null mutant of deltex; UAS-Mam, UAS-Flag Dx, UAS-NICD, and UAS-Dx;C96 GAL4 (kindly provided by Spyros Artavanis-Tsakonas, Harvard Medical School, Department of Cell Biology, Boston, MA); P {XP} d08059 (mahed08059) and EP1347 (maheEP1347) lines were used to study the loss-of-function effect of mahe [P {XP} d08059 (mahed08059) has an XP element inserted in the first intron upstream of the coding region of mahe and EP1347 (maheEP1347) has an EP element inserted in the first exon upstream of the coding region of mahe]; patched-GAL4: UAS-GFP (kindly provided by Florenci Serras, Institute of Biomedicine, University of Barcelona); Act5C–GAL4, eyeless-GAL4, apterous-GAL4, UAS-DIAP1, sgs-GAL4, patched-GAL4, and EP1347 were procured from Bloomington Stock Center; and P {XP} d08059 was from Exelixis Stock Center.
UAS-HA Mahe transgenic flies generation
To obtain full-length HA-tagged Mahe, the coding sequence of Mahe was amplified from full-length complementary DNA (cDNA) (GenBank accession no. AY060404.1) and was cloned in pUAST vector.
PCR amplification of the coding region of mahe was carried out using HA-tagged forward primer 5′-CGCAGGGTACCCAACATGTACCCATACGATGTTCCAGATTACGCTATGTTCGCTGGAGCATATGCACCC-3′and reverse primer 5′-GCGAGTCTAGATCACTACTCGATGGCCTGGCC-3′. The amplicon was digested with KpnI and XbaI, purified, and ligated in pUAST vector. Positive transformants were used for generating transgenic flies.
RNA extraction and semiquantitative RT-PCR
Total RNA was extracted from brain, eye-antennal discs, salivary glands, wing discs, fat body, and leg discs dissected from wandering third instar larvae, using the RNA extraction kit (Ambion). The extracted RNA was subjected to RNAase-free DNaseI treatment for 30 min at 37° to remove genomic DNA contamination. First-strand cDNA was synthesized using 2 µg of RNA as template, using 200 units of M-MulV reverse transcriptase (New England Biolabs, Beverly, MA) and 80 pmol of random primer (New England Biolabs).
Semiquantitative RT-PCR was performed with a mahe specific exon–exon junction primer. rps17 was used as an internal control:
mahe forward primer: 5′-TTCGTGCGTTGGCCCTTGTTATTG-3′
mahe reverse primer: 5′-GCTGGGCATCGAACGAGCAAG-3′
rps17 forward primer: 5′-AAGCGCATCTGCGAGGAG-3′
rps17 reverse primer: 5′-CCTCCTCCTGCAACTTGATG-3′.
Real-time PCR
The level of Notch expression was monitored using real-time, quantitative PCR. Reverse transcription was performed with a cDNA synthesis kit (Applied Biosystems, Foster City, CA), using aliquots of total RNA extracted. Real-time PCR reactions were performed using the ABI 7500 sequence detection system (Applied Biosystems) with SYBR Green PCR Master Mix (Thermo Scientific). The experiments were carried out in duplicate for each data set. The relative quantification in gene expression was determined using the 2−ΔΔCt method (Schmittgen and Livak 2008). The fold changes in gene expression were normalized to an internal control gene. rps17 was used as an internal control to normalize all data.
Primers used for the study are as follows:
Notch forward primer: 5′-AGCGAAATGGAGTCGGTCCCG-3′
Notch reverse primer: 5′-GATGGCGAGCCCAAGTAGGCA-3′.
Embryo collection
Embryos were collected on a 2% agar plate supplemented with 0.2% propionic acid and yeast paste. Embryos were washed with distilled water, dechorionated, and fixed for 1 hr in 1:1 heptane and 4% paraformaldehyde solution. Embryos were then devitellinized by replacing heptane with methanol followed by vigorous shaking. Devitellinized embryos were stored in methanol at −20°. Embryonic stages were identified as described in Campus-Ortega and Hartenstein (1985).
RNA–RNA in situ hybridization
To detect the pattern of transcript expression during embryogenesis, RNA in situ hybridization of mahe transcript was performed using a digoxygenin-labeled antisense mahe RNA probe. For RNA probe preparation, a 190-bp sequence of mahe was PCR amplified with mahe specific primers:
mahe forward primer: 5′-CGCAGAAGCTTGGACAACAATACGCTAACGCGC-3′
mahe reverse primer: 5′-GCGAGGAATTCGCAACTGTCGGGGTGTGTGTG-3′.
The PCR amplicon was double digested with HindIII and EcoRI followed by purification and ligation with HindIII- and EcoRI-digested pGEM-3Z vector. Positive clones were selected for large-scale plasmid extraction and generation of probe. The digoxygenin-labeled probe was synthesized using linearized pGEM-3Z-mahe construct following the manufacturer’s instructions (Roche). RNA–RNA in situ hybridization was carried out in embryos as described in Mutsuddi et al. (2004) and color detection was done with Sigma (St. Louis) FAST NBT/BCIP. Embryos were mounted in 1,4,diazabicyclo [2,2,2] octane (DABCO) and visualized in bright field under a Nikon (Garden City, NY) Eclipse 80i. Images were captured under suitable magnification and were processed with Adobe Photoshop 7.0.
Antibody generation and Western blotting
GST-Mahe fusion protein was used for generating the antibody. For synthesizing GST-Mahe fusion protein, the N-terminal region of Mahe was PCR amplified using forward primer 5′-CGCAGGAATTCATGTTCGCTGGAGCATATG-3′ with EcoRI and reverse primer 5′-GCGAGGCGGCCGCTTACAGTGTATTCGGATGAATG-3′ with NotI restriction sites. The PCR amplicon as well as pGEX-4T-1 (Amersham, Piscataway, NJ) plasmid was double digested with EcoRI and NotI, purified, ligated, and transformed into Escherichia coli BL21(GE Healthcare) cells for expression of GST-Mahe fusion protein. Protein expression was induced with 1 mM IPTG, purified on a glutathione sepharose column (GE Healthcare) per manufacturer’s instructions, and then used for raising polyclonal antibody in rabbit. Specificity of antibody was checked by performing Western blotting of protein extracted from wild-type as well as HA-tagged Mahe that was overexpressed in transformants. HA-Mahe expression was driven with a salivary gland-specific GAL4 driver (sgs-GAL4) and the protein sample was prepared from the salivary gland of third instar larvae. Cells were lysed in RIPA buffer (Cell Signaling) and were subjected to 10% SDS–PAGE followed by Western blotting. The blot was incubated with rabbit anti-Mahe (1:2000) or rabbit anti-HA antibody (1:1000; Sigma) or mouse anti-α-tubulin (1:1000; Sigma). Alkaline phosphatase (AP)-conjugated goat anti-rabbit, goat anti-mouse secondary antibody (Amersham; 1:2000) and color were developed with Sigma FAST NBT/BCIP. Three independent experiments showed similar results.
Immunocytochemistry and confocal microscopy
Immunostaining was performed in various tissues dissected from wandering third instar larvae. Larvae were dissected in cold PBS (pH 7.4) and immunostaining was done as described previously (Mukherjee et al. 2005). Staining with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (1µg/µl) was done to mark the nuclei. Tissues were mounted in DABCO. Images were examined with a Zeiss (Thornwood, NY) LSM 510 Meta laser scanning confocal microscope and processed with Adobe Photoshop7. Primary antibodies used were rabbit anti-Mahe, 1:200; mouse anti-Notch, 1:300 [C17.9C6; Developmental Studies Hybridoma Bank (DSHB)]; mouse anti-Cut, 1:100 (2B10; DSHB); mouse anti-Wg, 1:100 (4D4; DSHB); mouse anti-HA, 1:100 (Sigma); rabbit anti-HA, 1:100 (Sigma); and rabbit anti-Flag, 1:1000 (Sigma). Secondary antibodies used were goat anti-mouse antibody alexafluor-555 and goat anti-rabbit antibody alexafluor-555 (Molecular Probes, Eugene, OR) at a dilution of 1:200 and goat anti-rabbit antibodies conjugated with FITC at 1:100 dilution (Jackson ImmunoResearch Laboratories, West Grove, PA).
To monitor the levels of NECD, a detergent-free staining was performed using mouse anti-Notch extracellular domain-specific antibody (C458.2H, at 1:100 dilution; Developmental Studies Hybridoma Bank) with a similar protocol.
Data availability
Data available upon request.
Results
mahe encodes a putative DEAD box helicase that is conserved across taxa
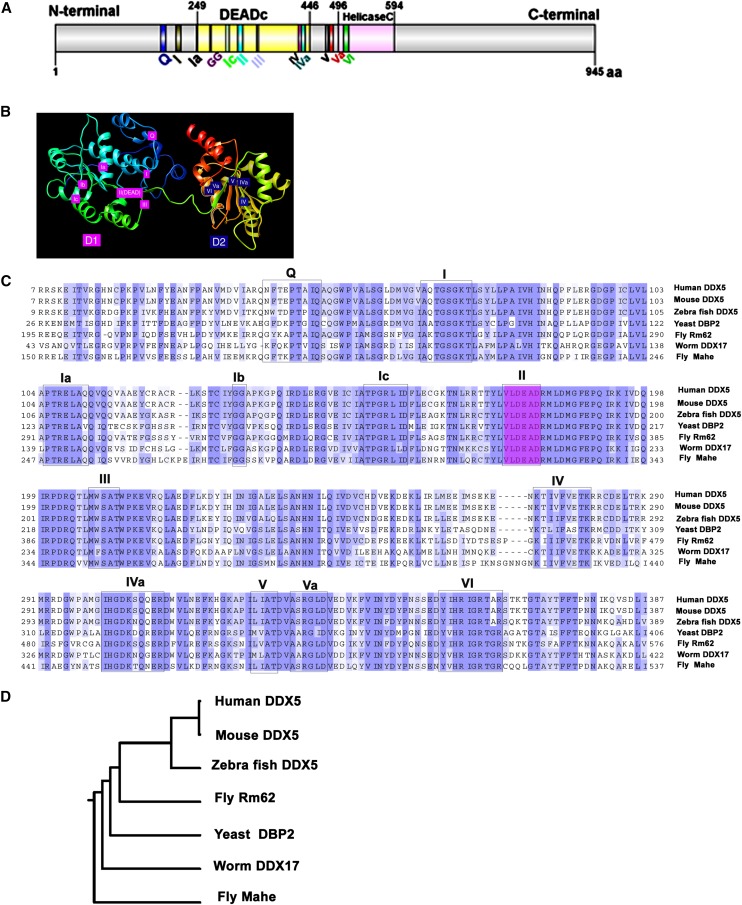
We have identified a novel DEAD box helicase mahe, which is predicted to comprise five exons and codes for a transcript that is ∼8.365 kb in size (accession no. NM-132196.3). It is located on the 7C3–7C4 region of the X chromosome (www.flybase.org). It encodes a putative RNA-binding protein that belongs to the highly conserved DEAD box helicase family, is 945 amino acids long (Figure 1, A and B), and is predicted to have a molecular weight of 110 kDa (http://web.expasy.org). Comparison of the deduced amino acid sequence of Mahe with ortholog prediction from the DRSC Integrative Ortholog Prediction Tool (DIOPT) shows that its ortholog is present in Homo sapiens, Mus musculus, Caenorhabditis elegans, Danio rerio, and Saccharomyces cerevisiae (Figure 1, C and D). Pairwise alignment of the Mahe amino acid sequence shows 54% and 53% identity with H. sapiens and S. cerevisiae, respectively (Figure 1C).
Figure 1.
mahe encodes a putative DEAD box RNA helicase that is highly conserved across the taxa. (A) Schematic representation of the predicted Mahe protein depicting the conserved domains along with the signature motif DEAD (motif II) that is a hallmark of this family of proteins. (B) A homology-based 3D model of Mahe was built with a Swiss model program (ExPASy) and the motifs were labeled using chimeras. The model shows the presence of two domains, D1 and D2, connected through a linker. Motifs Q, I, Ia, GG (Ib), Ic, II, and III represent D1 and motifs IV, IVa, V, Va, Vb, and VI constitute D2. From earlier reports it is known that D1 is essential for ATP binding and D2 is needed for substrate binding while D1 and D2 cooperate together to unwind the RNA duplex. (C) Clustal Omega alignment illustrates that Mahe is conserved across taxa and shares significant similarity with human, mouse, and zebra fish DDX5; yeast DBP2; worm DDX17; and Rm62 in fly. The 12 highly conserved motifs characteristics of DEAD box helicases are boxed and the signature motif DEAD for the DEAD box family is highlighted in magenta. (D) A phylogeny tree was generated using Clustal X, which is based on the evolutionary distance, and the horizontal lines indicate the evolutionary relationship among various species.
The putative Mahe protein contains a highly conserved signature motif DEAD, which is the hallmark of the DEAD box RNA helicase family. Swiss model 2.0 was used for prediction of different motifs across two domains (based on crystal structure of PRP5, which shows the highest alignment score with Mahe). Twelve highly conserved motifs constitute the two domains D1 and D2, which cooperate to perform the unwinding of target nucleic acids. D1 includes motifs Q, I, Ia, Ib, Ic, II (DEAD), and III, whereas motifs IV, IVa, V, Va, and VI make up the D2 domain (Figure 1B). Amino acid sequence comparison of DDX5 (human), DBP2 (yeast) and Mahe (fly) revealed a significant degree of similarity, indicating that all the members might have a conserved role to play in gene regulation.
A phylogenetic tree was generated to show the homology of Mahe within the species (Figure 1D). On the basis of the conserved domain, Mahe was found to be evolutionarily close to DDX5 (human, mouse, and zebra fish homologs), worm DDX17, yeast DBP2, and fly Rm62, thus revealing that it might have an important role to play across taxa.
Dynamic pattern of mahe expression during development
To examine the tissue-specific expression of mahe during development, total RNA was extracted from a wide variety of third instar larval tissue. Semiquantitative RT-PCR was performed using exon–exon junction primers specific to mahe and rps17 was used as an internal control. mahe transcripts were present in all the larval tissues examined including the brain, eye-antennal disc, salivary gland, wing disc, fat body, and leg disc (Supporting Information, Figure S1).
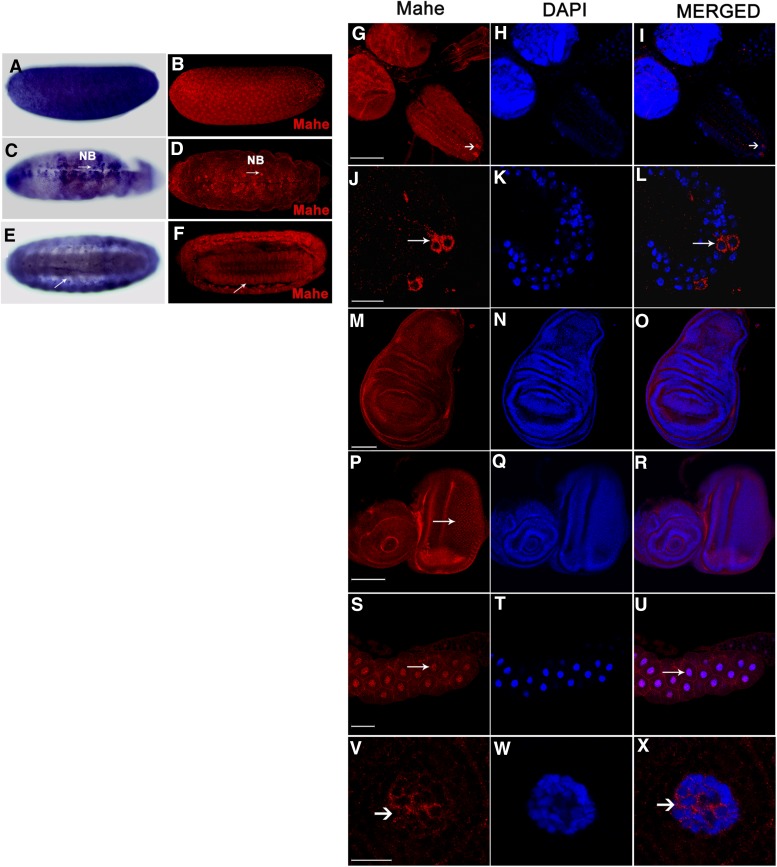
Whole-mount in situ hybridization was also carried out to detect the spatial pattern of expression of mahe during embryogenesis. Maternal deposit of mahe transcripts was observed during the early stages of embryogenesis in stages 1–3 (Figure 2A). A robust signal was seen in the neuroblasts from stage 11 embryos (Figure 2C) that are known to be the progenitor cells of the Drosophila nervous system, which eventually gives rise to neuronal and glial cells. A weak expression of mahe was also observed in the abdominal segments and the developing central nervous system (CNS) in stage 16 embryos (Figure 2E).
Figure 2.
Dynamic expression of mahe during Drosophila development. (A) RNA in situ hybridization shows maternal deposition of mahe transcript with strong localization in early-stage embryos. (C and E) Strong expression of mahe was also observed in the neuroblasts of stage 11 embryos (arrow) and in the central nervous system of late-stage embryos. (B, D, and F) Anti-Mahe was used to examine the pattern of expression of Mahe and DAPI was used to mark the nucleus. The embryonic expression of Mahe protein paralleled its transcript expression pattern (compare A, C, and E with B, D, and F). (G–X) Mahe expression was also observed in imaginal tissue of third instar larvae. A specific subset of neurons in the ventral nerve cord shows increased Mahe levels (arrow, G–I). This expression was confined to the cytoplasm of the neuronal cell body and was absent from the nucleus (arrow, J–L). Homogenous Mahe expression was found in the wing disc (M–O) along with expression in the ommatidia of the eye-antennal disc (arrow, P–R). A weak expression of Mahe was seen in the salivary gland nucleus as well as the cytoplasm (S–X), indicating tissue-specific subcellular localization of this protein. NB, neuroblast. Bars: G–I and M–U, 100 µm; J–L and V–X, 20 µm.
Polyclonal antibody was raised against Mahe protein (for details see Materials and Methods) and, as expected, a 110-kDa band was detected by Western blotting (Figure S2). This antibody was further used to examine the pattern of expression of mahe during development. Similar to that of the transcript expression, maternal accumulation as well as a strong localization of the protein was seen in the developing neuroblasts and the CNS (compare Figure 2, A, C, and E with Figure 2, B, D, and F).
Immunostaining with anti-Mahe in larval discs revealed strong Mahe localization in the optic lobes and the ventral nerve cord of the third instar larvae (Figure 2, G–L). In addition, Mahe protein was expressed at a higher level in a specific set of neurons in the ventral nerve cord (Figure 2, J–L). Homogenous expression of Mahe was observed in the wing discs (Figure 2, M–O), along with a strong expression in the photoreceptors of the eye-antennal discs (Figure 2, P–R). A weak, but detectable protein expression was observed in the salivary gland nucleus and cytoplasm (Figure 2, S–X). Taken together, a dynamic pattern of expression of mahe transcripts as well as protein was observed during development. The strong neuronal expression of mahe suggests its possible role during Drosophila neurogenesis.
Ectopic expression of mahe leads to phenotypes resembling that of Notch loss-of-function
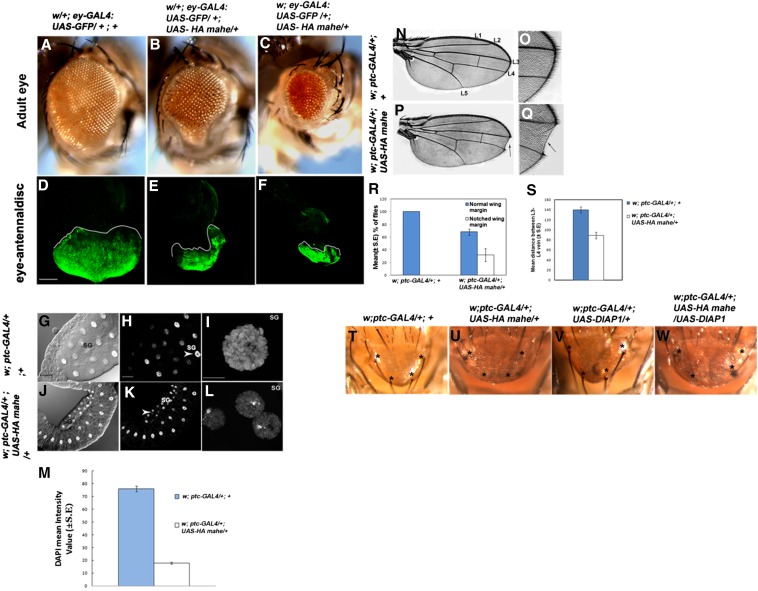
To gain insight into the precise function of mahe, the full-length coding sequence was subcloned along with an HA tag into pUAST vector. This construct included the DEAD and helicase domains of mahe. Overexpression of this putative RNA helicase using a variety of GAL4 drivers resulted in a wide range of phenotypes, such as lethality, notching of wings, and reduction in size of eye and salivary gland (Figure 3). The effects were dosage sensitive and increasing the copy number of the transgene led to worsening of the phenotype (data not shown). A variety of GAL4 driver lines were used to overexpress mahe and this resulted in a wide range of phenotypes (Table 1). Overexpression of mahe driven by Act5C-GAL4 and engrailed-GAL4 resulted in lethality at the second instar larval stage (Table 1). eyeless-GAL4 is known to be active in the eye and antennae anlagen by stage 11 of embryogenesis, which is followed by restricted expression only in the eye region of eye-antennal discs (Halder et al. 1998). Ectopic expression of mahe in eyeless-GAL4-driven cells resulted in a massive reduction in adult eye size in both male and female flies (100%, n = 234; Figure 3, A–F). The larval eye-antennal discs were also significantly smaller than those of the controls (n = 20; Figure 3, D–F). ptc-GAL4-driven expression of ectopic mahe in salivary gland also resulted in an enormous decrease in the size of the salivary gland with 100% penetrance (n = 10; Figure 3, G–L). In addition, the salivary gland nuclei from mahe overexpressed larvae (Figure 3, G–I) were much smaller than those of the controls (Figure 3, J–L); these nuclei were even smaller than the surrounding fat body nuclei, which served as an internal control since ptc-GAL4 does not drive HA-mahe in fat body nucleus. The total DNA content of the salivary gland nuclei was examined by staining with DAPI followed by quantification using the ZEN12 software package. Threefold reduction in DAPI staining was observed in the polytene nuclei with ectopically expressed mahe (Figure 3M).
Figure 3.
Ectopic expression of mahe results in development defects reminiscent of Notch loss-of-function. (A–C) Ectopic expression of mahe driven by ey-GAL4 induces a small eye phenotype in both female (B) and male (C) flies with 100% penetrance (n = 234). (D–F) Examination of the developing eye tissue of the eye-antennal disc (n = 20) (GFP fluorescence region marked with a dotted line) with ectopically expressed mahe shows significant reduction in size of tissues in both females (E) and males (F) (compare D to E and F). (G–L) ptc-GAL4-driven expression of mahe in salivary gland results in a dramatic reduction in size of salivary gland nuclei when compared to those of controls and the surrounding fat body nucleus with complete penetrance (n = 10). (M) The DAPI content of 10 salivary glands for both ptc-GAL4 >UAS-HA mahe and controls was quantified. DAPI content was found to be about three times less in mahe overexpressed salivary gland nuclei than in those of the controls. Values in M represent ±SE, P < 0.005. (N and O) Control adult wing displaying normal wing margin and longitudinal veins L1–L5. (P and Q) ptc-GAL4 >UAS-HA mahe wing shows notching at the wing margin (arrow) and reduced distance between L3 and L4 veins (black line), which is similar to that of loss of Notch signaling. (O and Q) Magnified inset of wild-type and ptc-GAL4>UAS-HA mahe wing shows that notching is confined to only the patched domain. (R) Quantification of flies with notching at the wing margin in ptc-GAL4 >UAS-HA mahe shows that >30% of flies have a notched wing margin (n = 223) while no notched wing is observed in control flies (n = 521). n indicates the total number of flies counted for each genotype and error bars represent standard deviation with P-value <0.0005. (S) Mean distance between L3 and L4 was calculated and it is observed that the distance between veins L3 and L4 is significantly reduced in ptc-GAL4 >UAS-HA-mahe. A total of 30 wings in three independent crosses were counted for both genotypes. Error bar represents ±SE and P-value <0.001. (T–W) Wild-type adult scutellum displays a regular arrangement and defined number of macrochaetae (asterisks, T) with externally observable socket and hair, while ptc-GAL4-driven mahe results in loss of hair as well as socket (asterisks, U). (V) Overexpression of an anti-apoptotic protein DIAP1 does not alter the bristle morphology. However, coexpression of DIAP1 (asterisk, W) and Mahe does not modify the balding phenotype induced by Mahe misexpression. n = total number of progeny. Bars: D–G and J, 100 µm; H, I, K, and L, 10 µm. SG, salivary gland; FB, fat body.
Table 1. Lethality and development defects induced by ectopic expression of mahe with a variety of GAL4 drivers.
| Genotype | Viability | Lethality | Adult phenotype |
|---|---|---|---|
| w;en-GAL4;UAS-HA mahe | Lethal | 8% embryonic lethality, 78% lethality at first instar, and 14% lethality at second instar (n = 364) | Lethal |
| w;Act5C-GAL4;UAS-HA mahe | Lethal | 40% lethality at embryonic level and 60% lethality at second instar stage (n = 688) | Lethal |
| w;patched-GAL4;UAS-HA mahe | Viable | ND | Viable with notched wing margin, reduced salivary gland size |
| w;eyGAL4:UAS-GFP;UAS-HA mahe | Viable | ND | Viable with smaller eye size and reduced salivary gland size |
| w;ap-GAL4;UAS-HA mahe | Viable | ND | Viable with blistered and notched wing |
ND, not determined; n, total number of embryos counted.
The effect of ectopic mahe expression in the developing wing imaginal discs was also examined. mahe expression at the anterior/posterior (A/P) boundary of the larval wing discs and scutellar region by the ptc-GAL4 driver led to notching of the wing margin (Figure 3, N–Q) along with reduced distance between the L3 and L4 veins (compare Figure 3N and 3P) as well as loss of sensory bristles in the scutellum (Figure 3U). These results suggest that an optimal level of Mahe is needed for proper development and differentiation in Drosophila and ectopic mahe expression phenocopies Notch loss-of-function.
It was observed that overexpression of mahe interferes with proper bristle formation in the scutellum, and a similar phenotype was also observed with Notch loss-of-function at the time of asymmetric division of the sensor organ precursor (SOP) cells (Hartenstein and Posakony 1990). Initially we thought that this bald phenotype may be because of apoptosis. However, anti-apoptotic protein DIAP1 could not suppress the mahe phenotype (compare Figure 3U and 3W), indicating that apoptosis was not the underlying cause of the balding phenotype. In the future, we need to dissect out the exact mechanism of the bald phenotype resulting from ectopic mahe expression to better understand the underlying molecular cause associated with this phenotype.
mahe genetically interacts with Notch and deltex alleles
Since ectopic expression of mahe leads to phenotypes resembling those of Notch loss-of-function, such as notching of wing margin and loss of scutellar bristles, we examined the genetic interaction of mahe with mutant alleles of genes involved in Notch signaling.
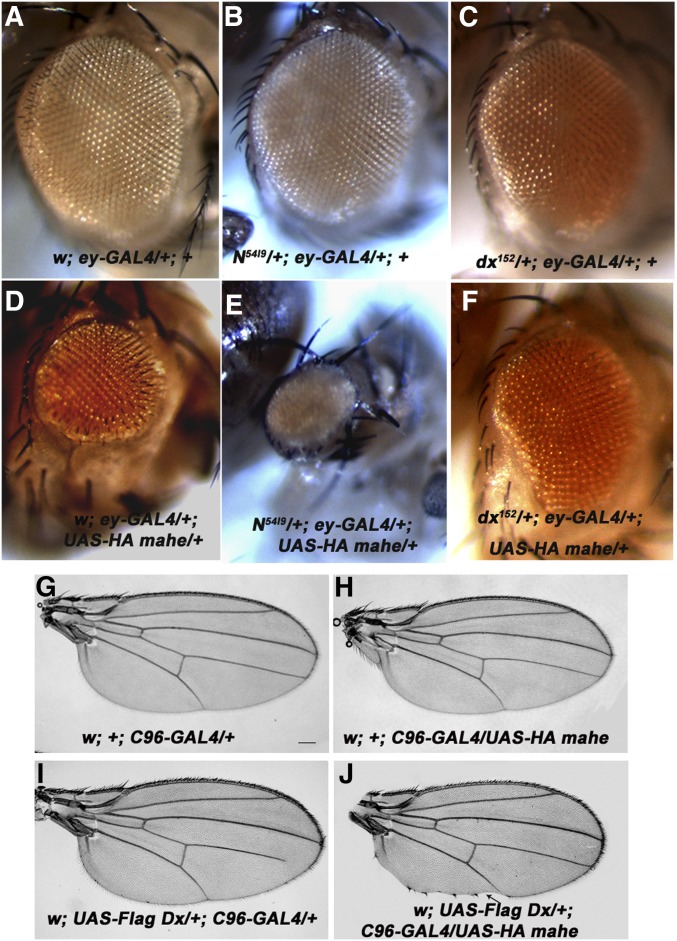
deltex encodes a cytoplasmic protein that is known to regulate Notch signaling both positively and negatively (Matsuno et al. 1995; Mukherjee et al. 2005). mahe overexpression was thus carried out in trans-heterozygous combination with N54l9, a null allele of Notch, and dx152, a loss-of-function allele of deltex. In heterozygous combination with N54l9, a mahe reduced eye phenotype was significantly enhanced with 100% penetrance (n = 12; compare Figure 4D and 4E), while dx152 marginally suppressed the reduced eye phenotype in 32% of the eyes examined (n = 29; Figure 4F). Further, the mutant alleles in combination with the eyeless-GAL4 driver had no effect on the eye size, clearly demonstrating that the genetic interactions of these alleles were with mahe and not with the GAL4 driver line (Figure 4, B and C). The effect of mahe overexpression on the Notch gain-of-function phenotype was also examined. Overexpression of mahe alone resulted in a massive reduction in the size of eye-antennal discs, which was completely penetrant (n = 20), while as reported earlier, expression of NICD alone resulted in highly proliferated eye tissue (Figure S4, B and C). Coexpression of Mahe along with NICD partially rescued the Notch-mediated proliferation in 6 of 20 eye antennal discs examined (Figure S4, D and H). Expression of Mahe alone with C96-GAL4 had no visible effect on the development of wing margin (Figure 4H), while expression of Dx alone with the C96-GAL4 driver resulted in mild irregularities of bristles at the wing margin (Figure 4I). However, coexpression of Mahe and Dx driven by C96-GAL4 resulted in nicking at the wing margin in 31% of the wings (n = 113) indicating downregulation of Notch function (Figure 4J). Additionally, genetic interaction of mahe was also checked with a nuclear protein Mam, which is a component of the coactivator complex of Notch signaling. Overexpression of a dominant-negative form of Mam at the wing margin resulted in severe serration of the wing margin (Figure S4I) and coexpression of mahe resulted in moderate enhancement of Mam-associated serration as well as a notching phenotype that was completely penetrant (n = 14; Figure S4J). It was important to examine genetic interaction of mahe loss-of-function/hypomorphic alleles in trans-heterozygous combination with Notch alleles as well as mutant alleles of genes functioning in Notch pathway.
Figure 4.
mahe genetically interacts with members of the Notch signaling pathway. (A) Adult eye with normal morphology. (B and C) N5419 and dx152 alone have no effect on eye size. (D) Overexpression of mahe with ey-GAL4 results in marked reduction in eye size in 100% of adults examined (n = 532). (E) Single copy of N5419, a null allele of Notch in combination with overexpressed mahe, results in a massive reduction in eye size in 100% of adult eyes examined (n = 12). (F) mahe overexpression in combination with dx152 (a null allele of deltex) results in mild rescue of eye size in 32% of adult eyes examined (n = 29). (G) Adult wing with C96-GAL4 driver alone shows normal morphology. (H) Expression of mahe with C96-GAL4 does not affect wing morphology. (I) Overexpression of Dx alone with C96-GAL4 results in mild irregularities of bristle pattern (n = 80). (J) Coexpression of Dx and Mahe results in wing nicking at the margin in 31% of wings examined (n = 113) (arrow). Bars in G–J, 100 µm.
We used two independent hypomorphic alleles of mahe, mahed08059 and maheEP1347, since mahe transcript levels were significantly lowered in these alleles (Figure S3A). Flies heterozygous for mahed08059 and maheEP1347 have normal adult wings (Figure S3, D and F). Flies heterozygous for N54l9 (Notch loss-of-function allele) have a low penetrant wing Notching phenotype (n = 28, 32%) (Figure S3B). A trans-heterozygous combination of N54l9 with either mahed08059 or maheEP1347 resulted in enhancement of the wing notching phenotype (n = 16, 50% and n = 23, 65%, respectively) (Figure S3, E and G).
Ectopic expression of mahe leads to loss of Cut and Wingless
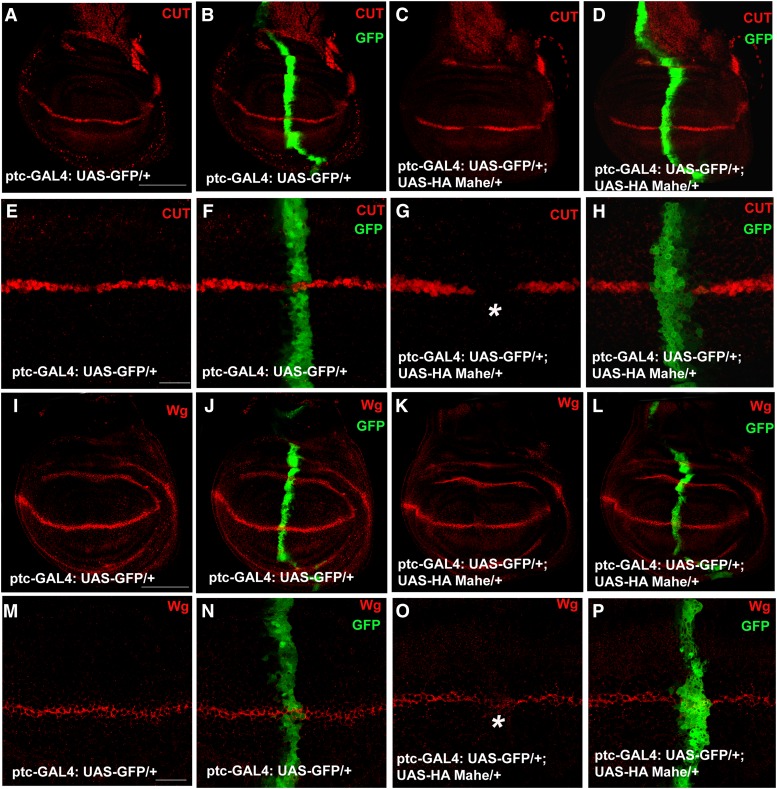
Notch signaling has been extensively studied in wing margin formation as well in SOP development. Proper expression of wingless (downstream target of Notch) requires active Notch signaling at the wing margin (Matsuno et al. 1995; Mukherjee et al. 2005). Similarly cut, another downstream target of Notch, requires active Notch signaling for its expression at the D/V boundary of the wing imaginal disc. Ectopic expression of mahe driven by ptc-GAL4 led to notching of the adult wing margin (Figure 3, P and Q). Since this phenotype was reminiscent of Notch loss-of-function, we monitored the expression of Cut and Wingless at the D/V boundary of the wing imaginal discs. In wild-type discs, Cut and Wingless are expressed in a few rows of cells at the D/V boundary (Figure 5, A, B, I, and J). Full-length HA-tagged Mahe overexpression at the A/P boundary of wing imaginal disc with ptc-GAL4 (marked with GFP) led to a complete loss of Cut and partial loss of Wingless at the D/V boundary (Figure 5, G and O) of the wing imaginal discs. The A/P boundary of the wing imaginal disc was marked with GFP in both the controls and ptc-GAL4-driven HA Mahe wing imaginal discs. Loss of Cut and Wingless was confined to GFP-marked cells, clearly revealing that loss of Cut and Wingless was specific to the cells in which mahe was overexpressed. Ectopically expressed HA-tagged Mahe was also checked by immunostaining with anti-HA antibody and reductions in both Cut and Wingless in the mahe expression domain were seen (Figure S5).
Figure 5.
Effect of overexpression of mahe on Cut and Wingless expression at the dorso-ventral boundary of the wing disc. (A–P) Cut and Wingless are the downstream targets of Notch and expressed in a narrow strip of cells at the dorso-ventral boundary of control wing discs as detected by anti-Cut (red) and anti-Wg (red). ptc-GAL4-driven expression of UAS-HA mahe at the anterior/posterior boundary of the wing disc (marked with GFP) results in complete loss of Cut (C, D, G, and H) and partial loss of Wingless expression (K, L, O, and P) at the dorso-ventral boundary of the wing disc (asterisks), indicating downregulation of Notch signaling. (H and P) Loss of Cut and Wg is specifically observed within the anterior/posterior boundary where mahe was overexpressed. Bars: A–D and I–L, 100 µm; E–H and M–P 20 µm.
Ectopic expression of mahe leads to reduction in Notch
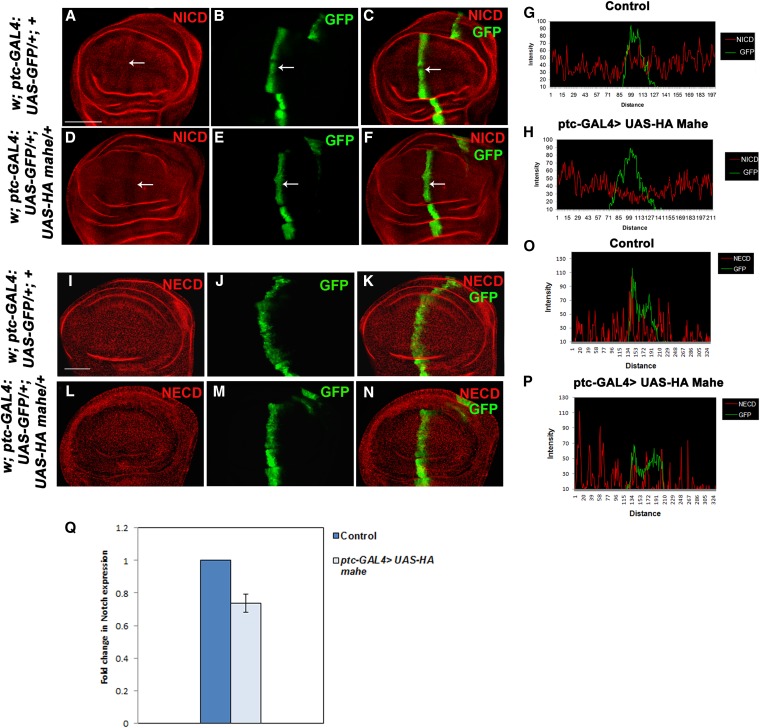
Since ectopic Mahe lowered the downstream targets of Notch signaling like Cut and Wingless, we examined whether Notch levels were also altered in these cells. The native Notch protein was detected with anti-Notch antibody raised against NICD, while ptc-GAL4-driven ectopic mahe-expressing cells were marked with GFP (Figure 6, A–H). Interestingly the GFP-marked cells with ectopically expressed Mahe exhibited significant reduction in endogenous Notch levels when compared to the control neighboring cells that were devoid of ectopic Mahe (Figure 6, D, F, and H). To check whether this reduction accounts for the depletion of full-length Notch protein, similar experiments were performed using antibody raised against NECD (Figure 6, I–P). However, no obvious change in NECD levels was observed upon ectopic expression of mahe (Figure 6, L, N, and P), suggesting that mahe overexpression does not affect full-length Notch receptor. Similarly, Notch transcript levels in cells with ectopic mahe also remained unaltered (Figure 6Q). However, the levels of Notch transcripts could not be examined in the wing discs since ptc-GAL4 drives expression in only a few rows of cells at the A/P boundary so instead we used eye-antennal disc extract for RNA isolation and monitored Notch transcript levels by real-time PCR with Notch-specific primers.
Figure 6.
Notch level was reduced upon mahe overexpression. (A–F) Notch protein was detected by an antibody generated against NICD epitope (red). (I–N) Surface Notch detected by an antibody against NECD epitope (red). (A–C) Normal distribution of Notch was seen at the anterior/posterior boundary (marked with GFP) in a control disc. (D–F) ptc-GAL4-driven expression of UAS-HA mahe at the anterior/posterior boundary of wing imaginal disc (marked with GFP) results in significant reduction of Notch protein in this domain (arrow) at 28°, suggesting a negative effect of Mahe on Notch levels. (G and H) Intensity profiling for NICD staining and GFP was generated using a confocal section for control and ptc-GAL4 >UAS-HA mahe wing imaginal disc. Note that the Notch levels were significantly reduced at the anterior/posterior boundary (marked with GFP) in ptc-GAL4 >UAS-HA mahe wing disc when compared to that of control disc. (I–K) Normal distribution of surface Notch as detected with NECD epitope was observed in a control disc. (L–N) ptc-GAL4-driven expression of UAS-HA mahe at the anterior/posterior boundary of wing imaginal disc (marked with GFP) did not alter Notch levels at the cell surface. (O and P) Intensity profiling for NECD staining and GFP shows that surface Notch levels were similar for control and ptc-GAL4 >UAS-HA mahe wing imaginal disc. Bars: A–F and I–N, 50 µm.
Coexpression of Mahe and Dx modulates Notch signaling
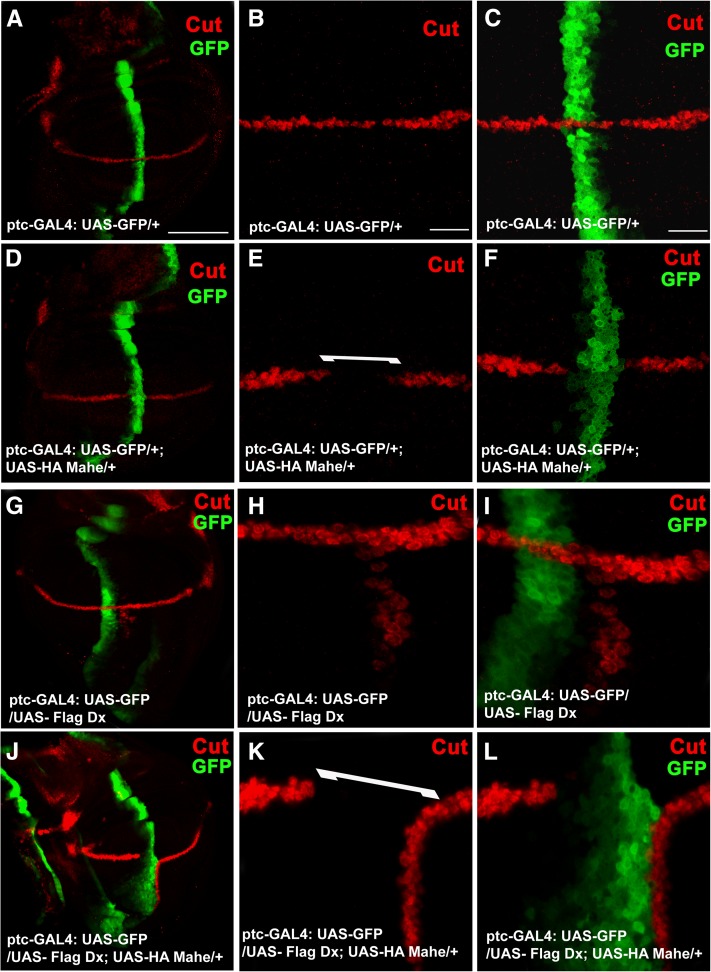
Since ectopic Mahe along with Dx results in a wing-nicking phenotype, we further went on to check the status of Notch signaling (monitored with Cut expression) in which both Dx and Mahe were coexpressed at the A/P boundary of wing discs. As mentioned earlier, overexpression of Mahe alone with ptc-GAL4 resulted in loss of Cut expression at the intersection of the A/P and D/V boundaries (Figure 7, D–F), while overexpression of Dx alone resulted in ectopic activation of Cut at the A/P boundary (Figure 7, G–I). Intriguingly, in combination with both overexpression of mahe and that of Dx, the loss of Cut was more pronounced at the intersection of the A/P and D/V boundaries (Figure 7, J–L). This clearly demonstrated that mahe in combination with Dx modulates Notch signaling.
Figure 7.
Mahe and Dx coexpression results in more pronounced loss of Cut expression at the D/V boundary. (A–C) Wing imaginal disc with normal Cut expression at the D/V boundary. (D–F) Overexpression of Mahe at the A/P boundary with ptc-GAL4 results in loss of Cut expression at the intersection of A/P and D/V boundaries. (G–I) Expression of Dx at the A/P boundary with ptc-GAL4 results in ectopic activation of Cut. (J–L) Coexpression of Mahe and Dx with ptc-GAL4 results in more severe loss of Cut at the intersection of A/P and D/V boundaries. Bars: A, D, G, and J, 100 µm; B, E, H, K, and C, F, I, L, 20 μm
Discussion
Notch signaling is an evolutionary conserved pathway required by a cell to communicate with its neighboring cells and regulates cell proliferation, cell differentiation, and cell death in metazoans (Artavanis-Tsakonas et al. 1983, 1999; Kopan 2002). The level of Notch signaling is an extremely important factor as its perturbation results in many developmental defects (Gridley 2003). Hence, fine-tuning of Notch signaling is vital, from its synthesis to its degradation (Artavanis-Tsakonas et al. 1995; Lai 2004) . Genome-wide studies have revealed a complex network of genetic circuits involved in context-dependent regulation of Notch signaling. There are multiple modifiers and regulators of the Notch signaling pathway, depending on the tissue type and developmental context, and more recently the involvement of RNA-binding proteins in regulation of Notch signaling has been appreciated (Hall et al. 2004; Saj et al. 2010; Jung et al. 2013).
We report characterization of a novel gene from the DEAD box family of RNA-binding protein, maheshvara (mahe), and have investigated its role in regulation of Notch signaling. Mahe belongs to the DEAD box family of protein as it harbors conserved motifs along with the signature motif DEAD. The pattern of expression of mahe was examined by in situ hybridization and immunostaining. Maternally accumulated mahe transcripts as well as protein are present in 0- to 2-hr embryos, followed by a strong expression in the embryonic neuroblast and larval CNS. In larval CNS, Mahe protein is accumulated in the cytoplasm of specific subsets of neurons. Overall, a strong neuronal enrichment of transcripts and protein was observed during embryonic development. Interestingly, overexpression of full-length protein with actin-GAL4 resulted in larval lethality, indicating that an optimum level of mahe is needed for cell survival. Ectopic expression of mahe also resulted in a wide variety of developmental defects, many of which were reminiscent of Notch loss-of-function. Notch signaling is involved in a wide variety of developmental processes and initially it was identified as a neurogenic gene, since loss-of-function alleles of Notch resulted in excess neurons (Artavanis-Tsakonas et al. 1995; Lai 2004). The enrichment of mahe transcripts and protein in the developing nervous system along with the loss of macrochaetae on the scutellum, which was associated with change in mahe levels, indicates that it may modulate the Notch-mediated cell fate specification. However, in the future we need to use markers for neuroblasts and study the role of mahe in neuronal cell specification.
The closest homolog of mahe in human, DDX5 (P68) has been reported to be involved in RNA splicing, transport, and stabilization as well as in microRNA biogenesis. However, recent studies by Jung et al. (2013) show that DDX5 physically interacts with MAML1 (Mam in Drosophila), which is a well-characterized member of the coactivator complex for Notch target genes. To understand the mechanism of how mahe regulates Notch signaling, we examined whether mahe genetically interacts with Mam. However, ectopic mahe marginally enhanced the serrate wing phenotype associated with Mam mutant, indicating that this novel RNA helicase may not be acting in the coactivator complex like that of the human counterpart, DDX5. We speculate that the mild enhancement of the Mam wing serration phenotype by mahe may be because of an additive effect, since both the genes might be interacting independently with Notch.
It is well established that classical Notch signaling is needed for cell fate determination during SOP development. Lateral inhibition mediated by Notch inhibits neighboring cells from adopting SOP fate; these SOPs then divide asymmetrically, leading to differential Notch distribution in a different population of cells, and ultimately affect cell fate determination. Loss of Notch function at the time of lateral inhibition results in the formation of supernumerary SOPs, while loss of Notch during asymmetric division causes cells to adopt a neuronal fate (Hartenstein and Posakony 1990). Our data do not support a reduction in Notch signaling during SOP formation, since downregulation of Notch signaling at this point will result in supernumerary SOP cells as a result of compromised lateral inhibition (Hartenstein and Posakony 1990; Posakony 1994). Instead, we speculate that ectopic Mahe leads to a reduction in Notch signaling at the time of asymmetric division with a change of cell fate from nonneuronal to supernumerary neurons, thus resulting in loss of bristles or a bald phenotype. However, a detailed genetic and molecular analysis is needed to show exactly how mahe interferes with Notch-mediated SOP development. To rule out that the bald phenotype caused by ectopic Mahe was not the result of apoptosis we checked whether this phenotype could be suppressed by an anti-apoptotic protein, DIAP1. DIAP1 overexpression did not alter the bald phenotype, suggesting that this phenotype might be an outcome of compromised Notch signaling and not because of apoptosis. However, ectopic mahe in wing imaginal discs induces mild cell death, which might correlate with involvement of mahe in regulation of another signaling pathway (S. Surabhi and M. Mutsuddi, unpublished data).
A remarkable decrease in nuclear size and DNA content of the salivary gland was also seen upon ectopic expression of mahe. A requirement of Notch signaling for endoreplication of ovarian follicle cells has been reported and a similar type of phenotype with reduced polyteny in salivary gland has been correlated with Notch loss-of-function (Deng et al. 2001; Martinez et al. 2009). Taken together these results suggest that mahe overexpression results in phenotypes that are reminiscent of loss of Notch signaling and further implicates the role of mahe in modulating Notch signaling at various levels during development.
One of the well-studied roles of Notch signaling is the establishment of the D/V boundary, where Notch maintains the expression of its targets Cut and Wg at the developing margin (Go et al. 1998; Micchelli et al. 1997). Loss of Cut and Wg at the D/V boundary of wing imaginal discs by ectopic mahe further supports the role of mahe in regulation of Notch signaling. Interestingly, we also observed significant reduction in NICD levels at the D/V boundary of wing imaginal discs upon mahe overexpression. In addition, we also report that depletion in NICD levels is seen while full-length Notch levels seem to be unaltered. Since mahe encodes a putative RNA-binding protein, we asked whether it directly regulates Notch transcript levels; however, no change in Notch transcripts was observed by modulating levels of Mahe. These data clearly indicate that Mahe regulates the level of processed Notch. We predict that the effect of Mahe on modulating the levels of NICD may be indirect, where Mahe might be regulating an important factor needed for maintenance of processed Notch.
Thus, our data suggest that although Mahe is an RNA helicase it does not alter Notch transcript levels. Since RNA-binding proteins are known to be key players in post-transcriptional gene regulation, it is possible that mahe regulates the turnover of NICD by affecting a crucial component of Notch signaling. A strong genetic interaction of Notch with both mahe gain-of-function and loss-of-function alleles further strengthens the involvement of mahe in regulation of Notch signaling. Strikingly, we also observed a strong genetic interaction of mahe with deltex in the wing as well as eye tissues. Coexpression of Dx and Mahe in the wing margin cells resulted in nicking at the wing margins that was absent with Dx or Mahe alone, indicating that Mahe along with Dx might negatively regulate Notch function.
Drosophila Dx is a cytoplasmic protein with a RING-H2, two WWE domains with an E3 ubiquitin ligase activity reported in mammalian homologs of Drosophila Dx (Takeyama et al. 2003). In Drosophila, Dx is known to regulate Notch signaling positively, as loss-of-function of deltex results in reduced Notch signaling (Matsuno et al. 1995). In mammals Deltex is known to antagonize Notch signaling (Izon et al. 2002; Kiaris et al. 2004). Additional studies postulate that Dx along with Kurtz, a nonvisual β-arrestin homolog in Drosophila, affects Notch signaling negatively in a ligand-independent manner (Mukherjee et al. 2005). Further, studies have also identified shrub, a core component in the ESCRT-III complex, as a modifier of Dx–Krz synergy and it has been proposed that shrub, Dx, and Kurtz regulate Notch trafficking and consequently its degradation in the late endosomal compartment (Hori et al. 2011). Hence two different roles in Notch regulation have been ascribed to Dx, depending on the tissue type and developmental context.
To understand the functional implication of Mahe and Deltex interaction, both Dx and Mahe were coexpressed at the A/P boundary of wing imaginal discs and Notch signaling output was monitored by immunostaining for Cut. Remarkably, loss of Cut expression at the D/V boundary by mahe was more pronounced in the Dx overexpressed background; based on these results we propose that along with Dx, mahe might play an important role in Notch regulation. In the future it will be interesting to dissect out the precise molecular mechanism of mahe and Dx interaction in the context of Notch regulation.
In conclusion we have identified a novel DEAD box helicase, mahe, which exhibits a very dynamic pattern of expression with significant enrichment in the developing CNS. Our studies show several lines of evidences that mahe regulates Notch signaling, which is known to be modulated at multiple steps. Notch signaling is also regulated at multiple steps. Our data explicitly present an interesting RNA helicase that regulates Notch signaling. In the future, we wish to further strengthen our observation by generating loss-of-function alleles of mahe. It is important to understand the precise mechanism of regulation of Notch signaling since perturbation of the Notch pathway results in inherited human diseases like Alagille syndrome, spondylocostal dysostosis, and cerebral autosomal dominant arteriopathy, etc. (Gridley 2003). Previously it was reported that RNA-binding proteins like Numb and Musashi regulate Notch signaling. Musashi activates Notch signaling by translational repression of Numb, which is a negative regulator of Notch (Rhyu et al. 1994; Imai et al. 2001). To this existing list we have added another gene mahe, which encodes an RNA-binding protein that is needed for proper maintenance of Notch signaling. Although the exact mechanism of this regulation is not known at this moment, in the future we wish to identify the candidate RNAs regulated by mahe, which will aid us in elucidating the molecular mechanism of how it regulates Notch signaling as well as other developmental processes.
Supplementary Material
Acknowledgments
We acknowledge the Bloomington Drosophila Stock Center and the Developmental Studies Hybridoma Bank at the University of Iowa for generously supplying stocks and reagents. We thank Florenci Serras and Spyros Artavanis-Tsakonas for kindly providing us with fly stocks. We acknowledge the confocal facility provided by the Department of Science and Technology, in the Department of Zoology and ISLS (Interdisciplinary Centre For Life Sciences), Banaras Hindu University. S.S. and B.K.T. were supported by a fellowship from the Council of Scientific and Industrial Research, Government of India. This work was supported by a grant from Department of Science and Technology, Government of India (to M.M.).
Footnotes
Communicating editor: P. Sengupta
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.181214/-/DC1.
Literature Cited
- Artavanis-Tsakonas S., Muskavitch M. A., Yedvobnick B., 1983. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 80(7): 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Matsuno K., Fortini M. E., 1995. Notch signalling. Science 268(5208): 225–232. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M. D., Lake R. J., 1999. Notch signalling: cell fate control and signal integration in development. Science 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Bond A. T., Mangus D. A., He F., Jacobson A., 2001. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol. Cell. Biol. 21: 7366–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C., Logeat F., Gupta N., Bessia C., LeBail O., et al. , 2000. A novel proteolytic cleavage involved in Notch signalling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5: 207–216. [DOI] [PubMed] [Google Scholar]
- Campus-Ortega J. A., Hartenstein V., 1985. Stages of Drosophila Embryogenesis: The Embryonic Development of Drosophila melanogaster. Springer-Verlag, Berlin, pp. 9–84. [Google Scholar]
- Caretti G., Lei E. P., Sartorelli V., 2007. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: eclectic regulators of disparate biological functions. Cell Cycle 6: 1172–1176. [DOI] [PubMed] [Google Scholar]
- Deng W. M., Althauser C., Ruohola-Baker H., 2001. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128: 4737–4746. [DOI] [PubMed] [Google Scholar]
- Go M. J., Eastman D. S., Artavanis-Tsakonas S., 1998. Cell proliferation control by Notch signaling in Drosophila development. Development 125: 2031–2040. [DOI] [PubMed] [Google Scholar]
- Gridley T., 2003. Notch signalling and inherited disease syndromes. Hum. Mol. Genet. 12 Spec No. 1: R9–R13. [DOI] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Flister S., Walldorf U., U. Kloter et al, 1998. Eyeless initiates the expression of both sineoculis and eyesabsent during Drosophila compound eye development. Development 125: 2181–2191. [DOI] [PubMed] [Google Scholar]
- Hall L. E., Alexander S. J., Chang M., Woodling N. S., Yedvobnick B., 2004. An EP overexpression screen for genetic modifiers of Notch pathway function in Drosophila melanogaster. Genet. Res. 83: 71–82. [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Posakony J. W., 1990. A dual function of the Notch gene in Drosophila sensillum development. Dev. Biol. 142: 13–30. [DOI] [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H., 1989. RNA helicase activity associated with the human p68 protein. Nature 339: 562–564. [DOI] [PubMed] [Google Scholar]
- Hori K., Sen A., Kirchhausen T., Artavanis-Tsakonas S., 2011. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J. Cell Biol. 195: 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., et al. , 2001. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 21(12): 3888–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izon D. J., Aster J. C., He Y., Weng A., Karnell F. G., et al. , 2002. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16: 231–243. [DOI] [PubMed] [Google Scholar]
- Jung C., Mittler G., Oswald F., Borggrefe T., 2013. RNA helicase Ddx5 and the noncoding RNA SRA act as coactivators in the Notch signaling pathway. Biochim. Biophys. Acta 1833: 1180–1189. [DOI] [PubMed] [Google Scholar]
- Kiaris H., Politi K., Grimm L. M., Szabolcs M., Fisher P., et al. , 2004. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am. J. Pathol. 165(2): 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., 2002. Notch: a membrane-bound transcription factor. J. Cell Sci. 115: 1095–1097. [DOI] [PubMed] [Google Scholar]
- Lai E. C., 2004. Notch signalling: control of cell communication and cell fate. Development 131(5): 965–973. [DOI] [PubMed] [Google Scholar]
- Lieber T., Kidd S., Young M. W., 2002. Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 16(2): 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., et al. , 1989. Birth of the D-E-A-D box. Nature 337: 121–122. [DOI] [PubMed] [Google Scholar]
- Liu F., Putnam A., Jankowsky E., 2008. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. USA 105: 20209–20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., et al. , 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95: 8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. M., Schuettengruber B., Sakr S., Janic A., Gonzalez C., et al. , 2009. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat. Genet. 41: 1076–1082. [DOI] [PubMed] [Google Scholar]
- Matsuno K., Diederich R. J., Go M. J., Blaumueller C. M., Artavanis-Tsakonas S., 1995. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121: 2633–2644. [DOI] [PubMed] [Google Scholar]
- Micchelli C. A., Rulifson E. J., Blair S. S., 1997. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124(8): 1485–1495. [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Veraksa A., Bauer A., Rosse C., J. Camonis et al, 2005. Regulation of Notch signalling by non-visual beta-arrestin. Nat. Cell Biol. 7: 1191–1201. [DOI] [PubMed] [Google Scholar]
- Mutsuddi M., Marshall C. M., Benzow K. A., Koob M. D., Rebay I., 2004. The spinocerebellar ataxia 8 noncoding RNA causes neurodegeneration and associates with staufen in Drosophila. Curr. Biol. 14: 302–308. [DOI] [PubMed] [Google Scholar]
- Posakony J. W., 1994. Nature vs. nurture: asymmetric cell divisions in Drosophila bristle development. Cell 76: 415–418. [DOI] [PubMed] [Google Scholar]
- Rhyu M. S., Jan L. Y., Jan Y. N., 1994. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76: 477–491. [DOI] [PubMed] [Google Scholar]
- Rocak S., Linder P., 2004. DEAD-BOX protein: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell. Biol. 5: 232–241. [DOI] [PubMed] [Google Scholar]
- Saj A., Z. Arziman, D. Stempfle, W. V. Belle, U. Sauder et al., 2010 A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev. Cell 18: 862–876. [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J., 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 6: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Sengoku T., Nureki O., Dohmae N., Nakamura A., Yokoyama S., 2004. Crystallization and preliminary X-ray analysis of the helicase domains of Vasa complexed with RNA and an ATP analogue. Acta Crystallogr. D Biol. Crystallogr. 60: 320–322. [DOI] [PubMed] [Google Scholar]
- Struhl G., Greenwald I., 1999. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398: 522–525. [DOI] [PubMed] [Google Scholar]
- Takeyama K., Aguiar R. C. T., Gu L., He C., Freeman G. J., et al. , 2003. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem. 278: 21930–21937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available upon request.