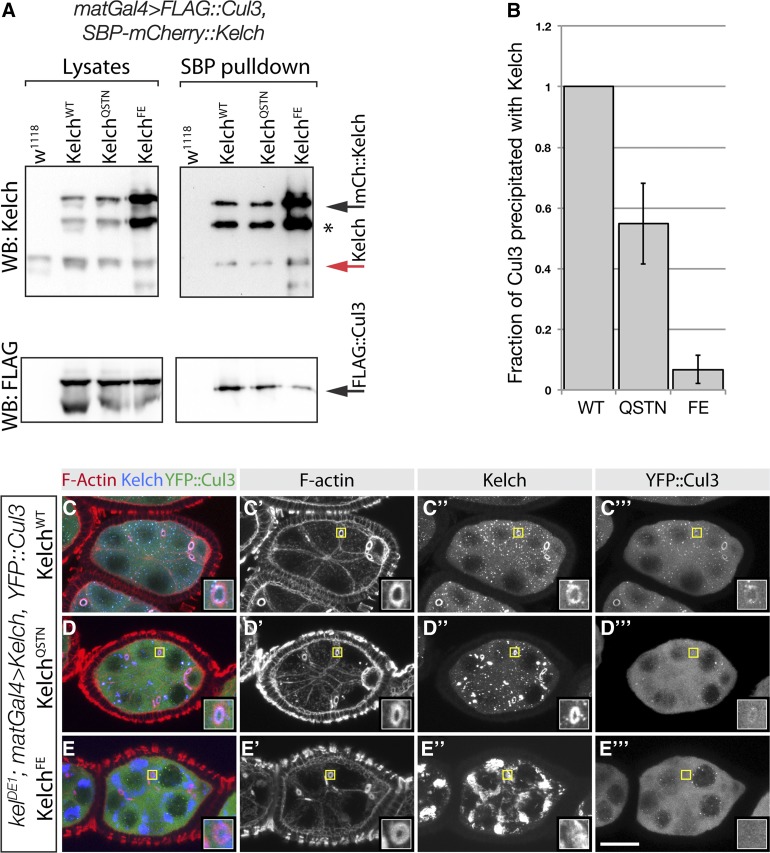
Figure 5.
Mutations in the Kelch BTB domain disrupt the association of Kelch with Cul3 in vivo. (A) Western analysis of pulldown experiment to analyze complex formation between Cul3 and Kelch BTB mutants. Ovarian lysates were prepared from flies expressing SBP–mCherry-tagged Kelch proteins in a wild-type background and subjected to pulldowns with Streptavidin beads. Western blots of pulldowns and input lysates were probed with Kel 1B and FLAG M2 monoclonal antibodies. Red arrow indicates endogenous Kelch, the presence of which in the pulldown experiments is an indication of dimerization of endogenous Kelch with the tagged Kelch protein. Asterisk indicates a proteolytic product of the overexpressed SBP–mCherry::Kelch proteins. Black arrow indicates full-length SBP-mCherry::Kelch. (B) Analysis of Cul3 coprecipitation efficiency. Cul3 precipitation efficiency is plotted as a ratio of the amount of Cul3 precipitated to the amount of Kelch in the Streptavidin precipitates. The amount of Kelch and Cul3 precipitated from flies expressing tagged SBP–mCherry::KelchWT was set at 1. Error bars indicate the standard error of the mean from three independent experiments. The ratios obtained with each Kelch protein differed significantly from the other two (P < 0.05, one-way ANOVA, Tukey’s multiple comparison test). (C–E) Egg chambers from homozygous kelDE1 flies expressing the indicated Kelch protein and Venus::Cul3. Regions indicated by yellow boxes are shown as higher magnification insets. YFP::Cul3 localized to kelDE1 ring canals rescued by wild-type Kelch (C–C′′′) and KelchQSTN (D–D′′′), but not KelchFE (E–E′′′).