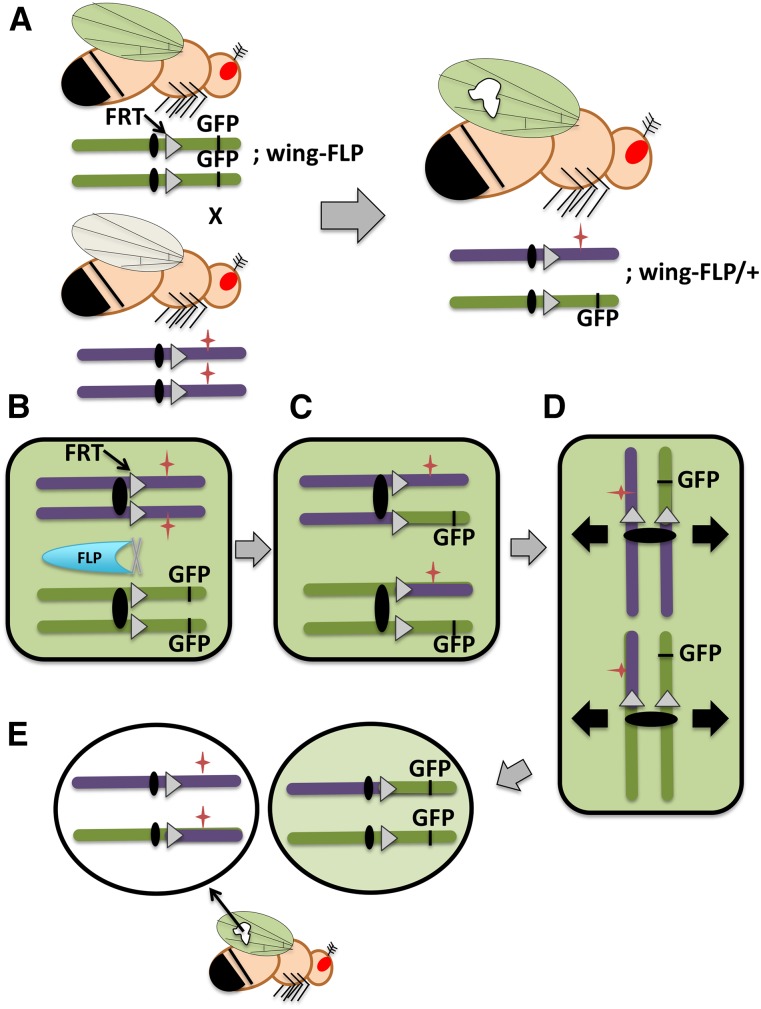
Figure 6.
Clonal analysis in somatic tissue. (A) Schematic diagram of a genetic cross used to create homozygous somatic clones of a mutation of interest within a heterozygous background. In this example, the clone is a patch of mutant cells within a wild-type wing. One parent is homozygous for a chromosome that carries the FLP recombinase recognition site, FRT (triangles) and a distal GFP marker that is being expressed under the control of a wing promoter. All of the cells of this fly’s wings will express GFP and appear green under a fluorescent microscope. This fly is also homozygous on another chromosome (not drawn) for the FLP recombinase gene, which is being expressed under the control of a wing promoter. The other parental fly is homozygous for a mutation of interest (red star) on the same FRT-carrying chromosome. Progeny from this cross will be transheterozygous for the GFP-marked chromosome and the mutant chromosome, and they will be heterozygous for FLP. During development, FLP-mediated mitotic recombination in the developing wing will produce patches of unmarked homozygous mutant cells (white patch). Panels B–E show the mechanics of clone production through mitotic recombination in the progeny. (B) Cells in mitotic G2 have replicated chromosomes with sister chromatids. In some wing cells, the FLP recombinase triggers recombination between FRT sites on nonsister chromatids, and (C) one copy each of the GFP marker and the mutation of interest will switch between homologous chromosomes. (D) One of two possible chromatid alignments at mitotic metaphase for the cell pictured in C. (The other alignment, not shown, leads to two heterozygous daughter cells.) The black arrows show the direction of sister chromatid separation during the completion of mitotic division. (E) Daughter cells produced upon completion of cell division for the cell pictured in D. One daughter cell is homozygous for the mutation of interest and will subsequently divide to give rise to a homozygous patch of cells, which can be identified for phenotypic analysis based on loss of the GFP marker. The other daughter cell is homozygous for the GFP marker and will blend into the surrounding heterozygous cells that also fluoresce.