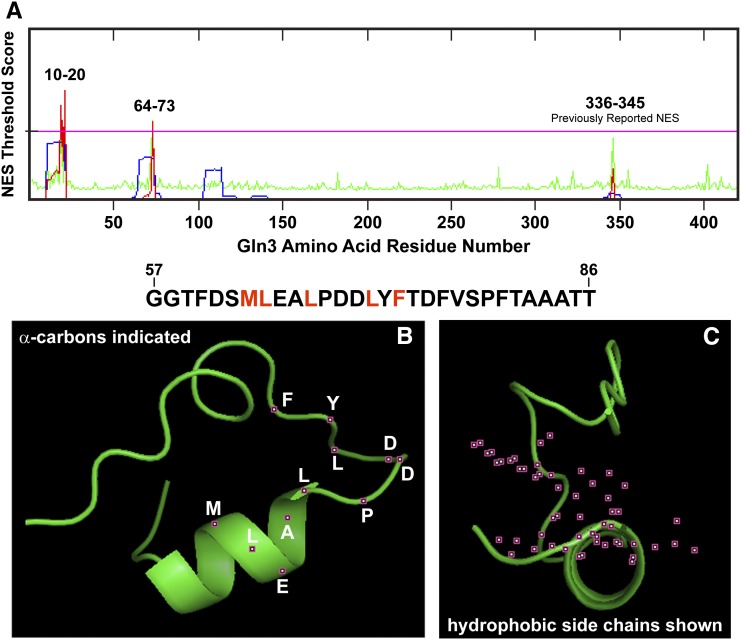
Figure 3.
Three Gln3 sequences yielded threshold prediction scores indicating their likelihood of functioning as NES motifs. The computer programs used to generate these outputs are described in the text. (A) The prediction server calculated the overall NES score (red trace) from the hidden Markov model (HMM, blue trace) and artificial neural network (ANN, green trace) scores. If the NES scores of individual residues exceed the threshold value (pink horizontal line), they are predicted to potentially function as a nuclear export signal. (B and C) The predicted most likely structure of Gln3 residues 57–86 (sequence appears above the graphics). Positions of the α-carbon atoms of the residues (white lettering) are indicated in the left image as small maroon boxes with white centers (B). The side-chain atoms of the critical hydrophobic residues in a canonical NES sequence (indicated in red in the sequence above the images) are indicated as small maroon boxes with white centers (C).