Fig. 3.
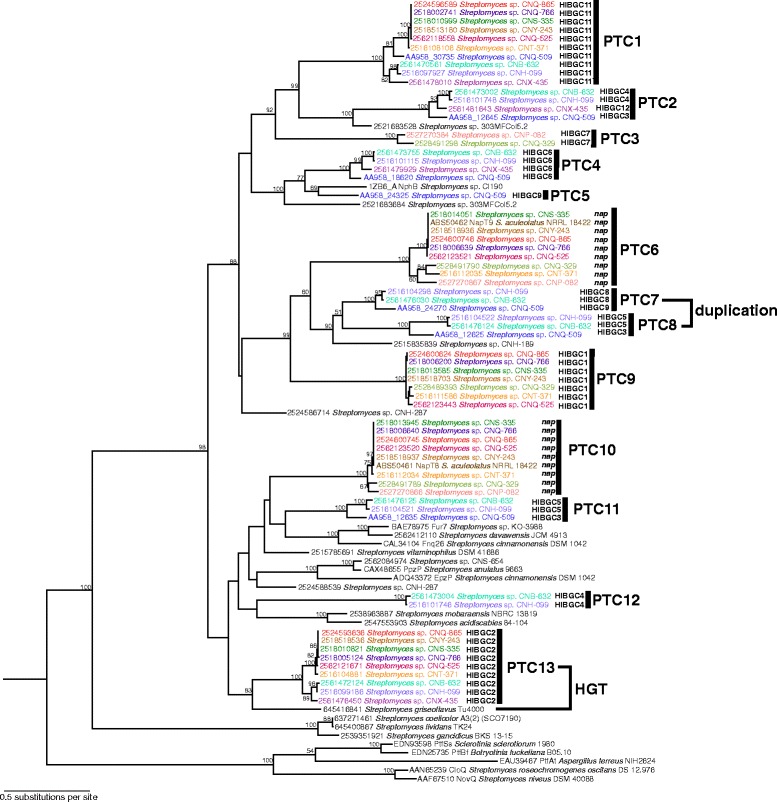
Maximum likelihood phylogeny of Orf2 ABBA PTases with mid-point rooting. The phylogeny contains all of the Orf2 ABBA PTases identified in the 120 Streptomyces genomes (identified by IMG gene number) including 12 that are experimentally characterized (identified by accession number). Homologs found in non-MAR4 genomes are in black, while those from MAR4 strains are color-coded based on strain. MAR4 PTases are delineated into 13 prenyltransferase clades (PTCs). Each MAR4 PTase is also assigned a hybrid isoprenoid gene cluster (HIBGC) number, which defines the gene cluster in which it was observed. Bootstrap values >50 % are indicated at their respective nodes (based on 100 replicates). Examples of horizontal gene transfer (HGT) and gene duplication are indicated. Two of the characterized PTases (ABS50462 and ABS50461) are from the MAR4 strain S. aculeolatus NRRL 18422
