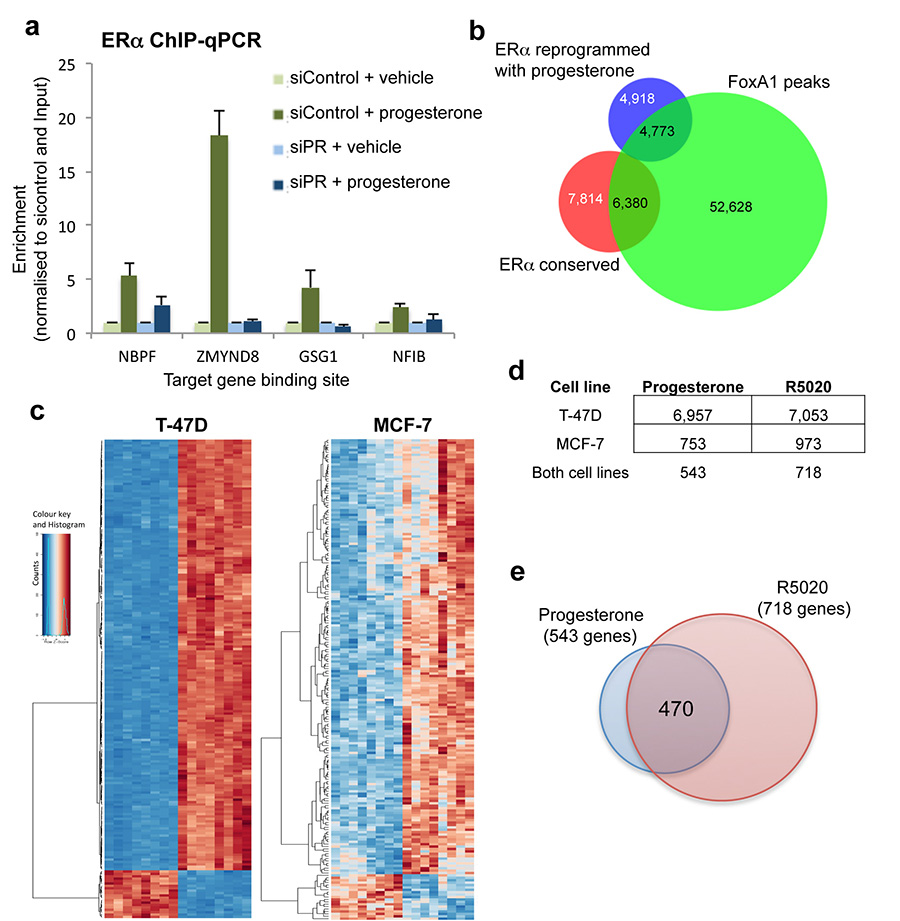
Extended data figure 4. Validation of binding and gene expression changes.
a. Validation of dependence on PR for ERα binding and overlap between ERα binding and FoxA1 binding. T-47D cells were grown in full, estrogen-rich media and transfected with siControl or siRNA to PR. ERα ChIP was conducted followed by qPCR of several novel ERα binding events only observed under progesterone treatment conditions. In the absence of PR, ERα is not able to associate with the progesterone-induced binding sites. The figure represents one biological replicate of three competed replicates and the error bars represent standard deviation of the technical ChIP-PCR replicates. b. Venn diagram showing the ERα binding events that were conserved in T-47D cells (i.e. not altered by progesterone when compared to estrogen alone) and the ERα binding events that were reprogrammed by progesterone treatment, when overlapped with FoxA1 ChIP-seq data from T-47D cells. The FoxA1 ChIP-seq data from T-47D cells was from Hurtado, et al, Nature Genetics, 2011, 43:27-33. c. Differential gene changes in MCF-7 and T-47D cells following treatment with progesterone or R5020 for 3hr. Heatmap showing gene changes relative to matched controls. Eight replicates were included. d. Table showing the differentially regulated genes in the two cell lines and in the two treatment conditions. e. Overlap between genes regulated by progesterone (in both cell lines) and gene regulated by the synthetic progestin R5020 (in both cell lines).