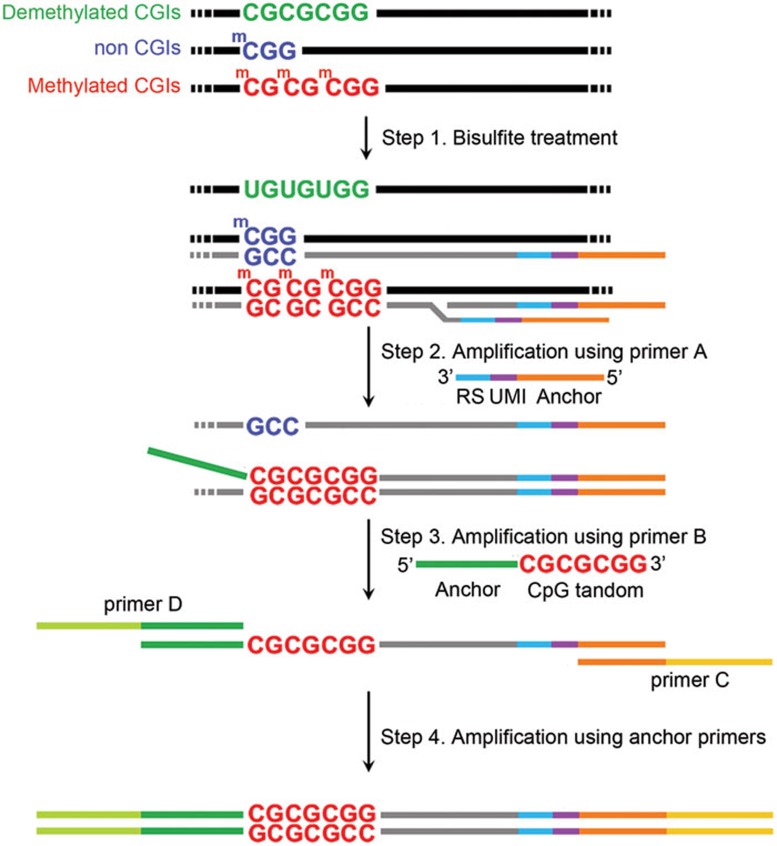
Figure 1.
Schematic of MCTA-Seq. First, the CpG tandems in the unmethylated CGIs are converted to UpG tandems via bisulfite treatment, whereas those in the methylated CGIs are unaffected. Second, the MCTA-Seq primer A binds semi-randomly to the converted DNA at the CpG site and is extended by a polymerase with displacement activity. The MCTA-Seq primer A consists of the following three parts: a semi-random sequence (RS) containing one CpG site (see Materials and Methods) is at the 3′-end, a unique molecular identifier (UMI) sequence is in the middle, and an anchor sequence is at the 5′-end. The methylated CGIs are expected to be amplified to a higher degree since they have high density of methylated CpG sites. Third, the MCTA-Seq primer B, which contains the CpG tandem sequence “CGCGCGG” at the 3′-end followed by a 4-bp “DDDD” sequence (D represents A or T or G), selectively amplifies the methylated CpG tandem sites. Last, exponential PCR amplification is performed using primer C and primer D, which is indexed, against the anchor sequences.