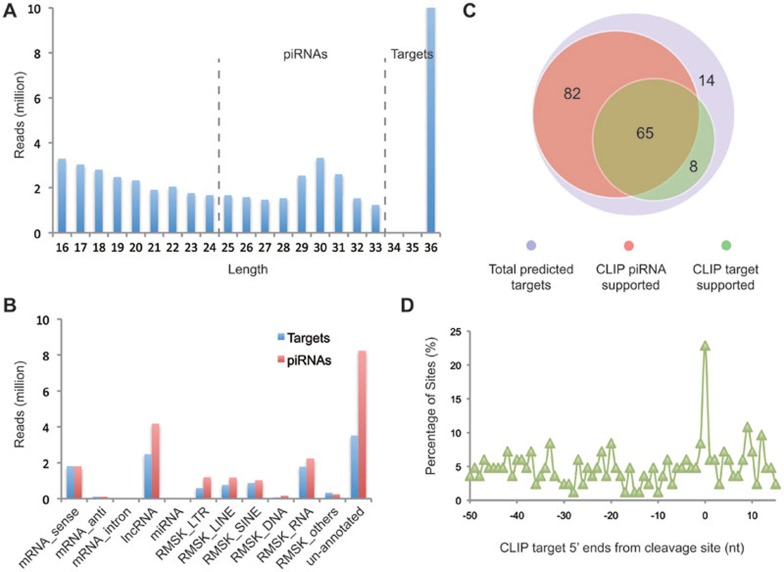
Figure 3.
piRNAs and piRNA targets identified by MIWI CLIP-seq. (A) The length distribution of mapped RNAs detected by MIWI CLIP-seq. Short sequences of 25-33 nt likely correspond to piRNAs, while longer sequences (only 36 nt were sequenced) correspond to MIWI targets. (B) Genome annotations for MIWI CLIP-identified targets and piRNAs. (C) Overlaps between genes that contain target sites bound by MIWI CLIP-identified piRNAs (red) and genes that contain target sites directly bound by MIWI (green) in 169 piRNA target genes (blue) predicted from the 5′-RACE library. Only the MIWI CLIP-identified piRNAs bound at the expected cleavage sites and the MIWI CLIP-identified mRNA sequences fallen within 15 nt from the cleavage sites were taken into account. (D) Alignment of the 5′ ends of MIWI CLIP-identified mRNA targets (> 33 nt) with the predicted cleavage sites from the 5′-RACE library. The Y-axis represents the percentage of predicted target sites with the existence of 5′ ends of MIWI CLIP-identified targets mapped at positions, as indicated by the X-axis, relative to the predicted cleavage sites.