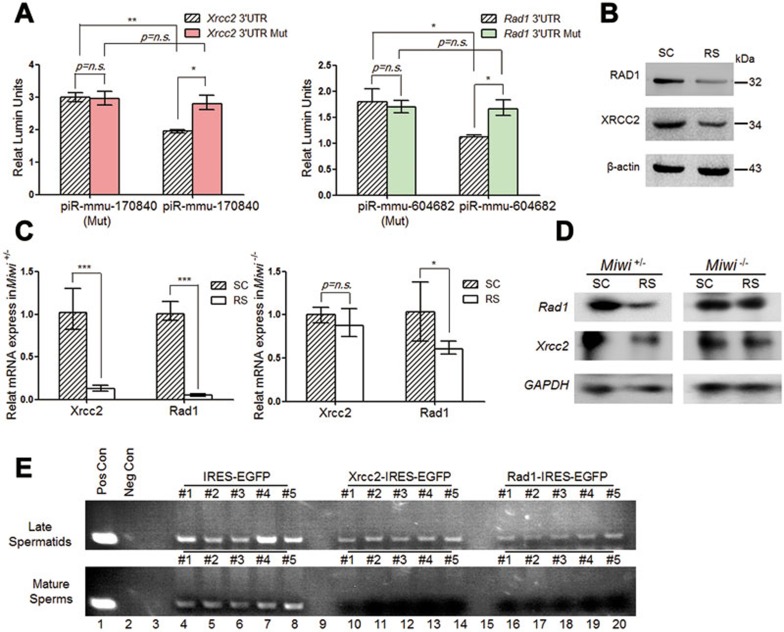
Figure 6.
Temporal regulation of the piRNA target genes Xrcc2 and Rad1 is essential for sperm formation. (A) Dual-luciferase reporter assays to determine the effects of piR-mmu-17084024 on the Xrcc2 reporter (left), and piR-mmu-60468224 on the Rad1 reporter (right) in transfected GC-2spd(ts) cells. A mutant piRNA (Mut) served as negative control in each case. (B) Western blot analyses of Xrcc2 and Rad1 expression in enriched SC and RS, with β-actin serving as the loading control. (C, D) qRT-PCR (C) or northern blot (D) analyses of the mRNA levels of Xrcc2 and Rad1 in Miwi+/− and Miwi−/− mouse spermatogenic cells, with GAPDH serving as an internal control. Mean ± SEM of three separate experiments were plotted. Student's t-test significance: *P < 0.05, **P < 0.01, ***P < 0.001. (E) PCR analyses of GFP DNA in extracted genomic DNA from late spermatids (top) and epididymal sperms (bottom) from mouse testes transduced with IRES-EGFP (lanes 4-8), Xrcc2-IRES-EGFP (lanes 10-14), or Rad1-IRES-EGFP (lanes 16-20), respectively. Genomic DNA samples from Oct4-GFP transgenic mice (lane 1) and wild-type background mice (lane 2) served as positive and negative controls, respectively. Results shown are representative of three independent experiments.