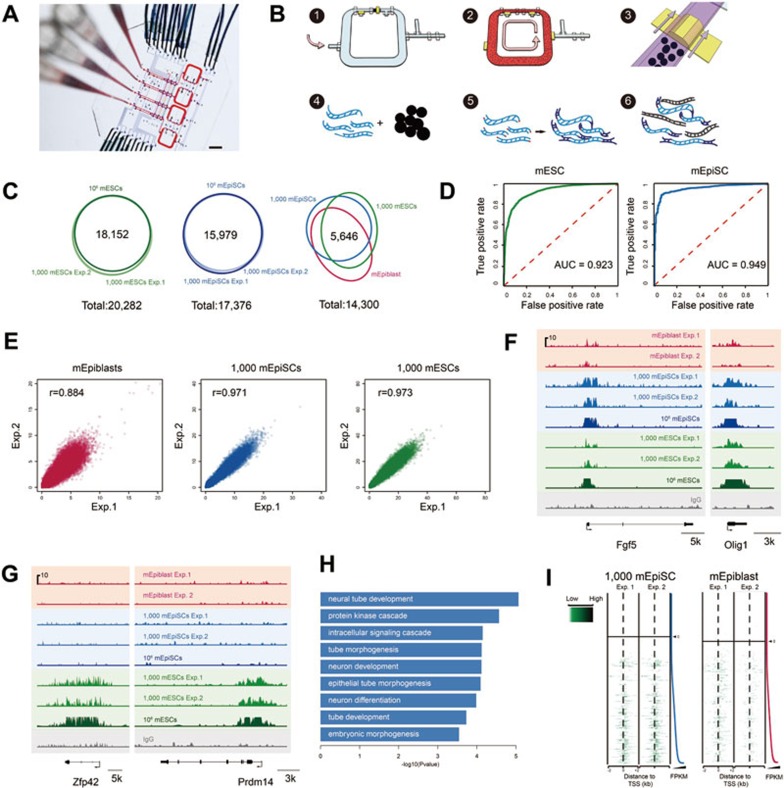
Figure 1.
Microfluidic device and the ChIP-Seq from 1 000 mEpiSCs, 1 000 mESCs and epiblast cells of the E6.5 mouse embryos. (A) Optical micrograph of a 4-plex microfluidic device with control lines and sample inputs. Scale bar: 5 mm. (B) Key operational steps of a ChIP-Seq flow pipeline. Step 1: load the chromatin fragments to fill dead-end flow-channels; Step 2: mixing and immunoprecipitation; Step 3: trap the antibody-functionalized beads (Ab-beads) to form a column; Step 4: release the DNA from the chromatin-antibody-bead complex; Step 5: end-repair, adenylation, and ligation; Step 6: amplification for sequencing. (C) The Venn diagram of enriched TSS regions of mEpiSCs, mESCs, and E6.5 epiblast cells. (D) The receiver operating characteristic (ROC) curve representing the true positive and false positive rates for the 1 000-cell experiment of mEpiSCs and mESCs. The standards are the enriched TSS regions called from the ChIP-Seq experiments using one million cells. (E) The correlation of the enrichment of H3K4me3 markers around TSS regions of epiblast cells of E6.5 mouse embryos, mEpiSCs, and mESCs. Each point represents an individual gene. (F) Representative loci of the ChIP-Seq of the H3K4me3 markers for three cell types, E6.5 epiblast cells, mEpiSCs, and mESCs, showing the peaks shared by all these three types of pluripotent cells. (G) Representative loci of the ChIP-Seq of the H3K4me3 markers specifically enriched in mESCs. (H) Gene ontology terms enriched in epiblast cells of E6.5 mouse embryos and mEpiSCs, compared to mESCs. (I) Heatmap showing read distribution around TSS regions of different transcripts ranked by the FPKM.