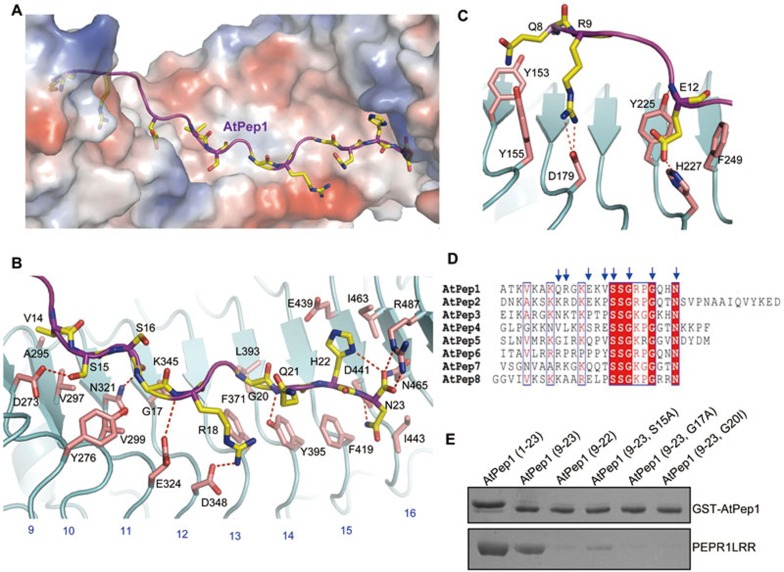
Figure 3.
Structural basis for recognition of AtPep1 by PEPR1LRR. (A) AtPep1 binds to a surface groove at the inner side of the PEPR1LRR solenoid. PEPR1LRR is shown in electrostatic surface and AtPep1 in cartoon. White, blue and red indicate neutral, positive and negative surfaces, respectively. The side chains of some amino acids from AtPep1 are shown (yellow and stick). (B) Interaction of the C-terminal portion (residues 14-23) of AtPep1 with PEPR1LRR. The side chains of PEPR1LRR and AtPep1 are shown in pink and yellow, respectively. Red dashed lines indicate hydrogen or salt bonds. Numbers in blue indicate the positions of LRRs. (C) Interaction of the N-terminal portion (residues 7-13) of AtPep1 with PEPR1LRR. (D) Sequence alignment of AtPeps from Arabidopsis. The conserved residues are shown with red background. Amino acids with arrows on the top are involved in interaction with PEPR1LRR. (E) Mutagenesis analysis of AtPep1 interaction with PEPR1LRR. The assay was performed as described in Figure 1A.