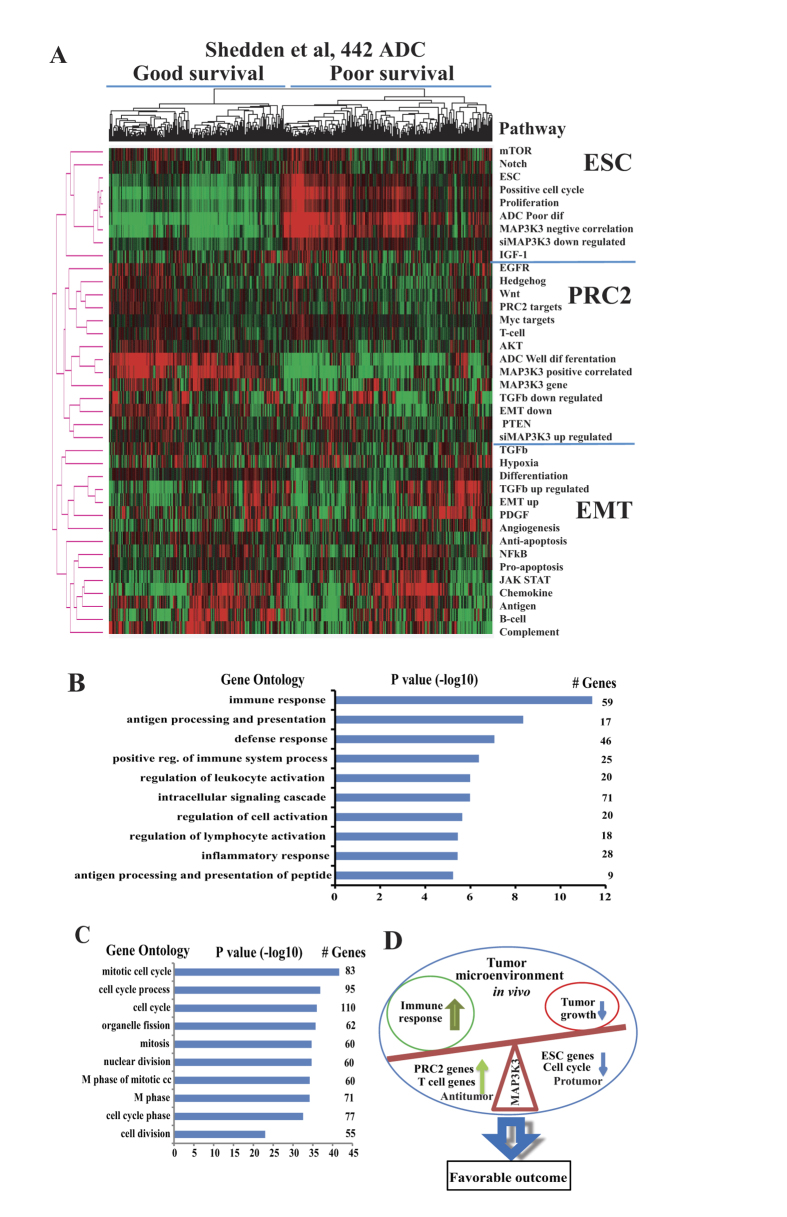
Figure 6. Cluster and DAVID pathway analysis of MAP3K3 mRNA correlated genes.
(A): Heat maps showing the unsupervised cluster analysis of MAP3K3 correlated (positive or negatively, based on Shedden et al., 442 ADCs by Spearman correlation) genes, MAP3K3 regulated (after MAP3K3 siRNA knockdown of H1299 and H838 cells) genes, most cancer related pathways, ESC (embryonic stem cell), PRC2 (polycomb repressive complex 2) and EMT (epithelial-mesenchymal transition) related gene signatures (mean expression value of each pathway or gene group was used). Three major clusters were found, ESC, PRC2 and EMT, based on their mRNA expression patterns. X-axis represents samples, y-axis pathways/MAP3K3 gene, red represents high expression and green low expression, (B): DAVID pathway or Go Ontology analysis of these MAP3K3 correlated genes indicates that the MAP3K3 positive correlated genes were significantly involved in immune response (p > 10 of –log10 value), and (C): MAP3K3 negatively correlated genes were significantly involved in cell cycle related process (p > 30 of –log10 value. (D): Potential working model for MAP3K3 in the tumor microenvironment of lung cancer. MAP3K3 overexpression may be beneficial for patient survival due to the change of balance between active immune response and tumor growth in the tumor environment.