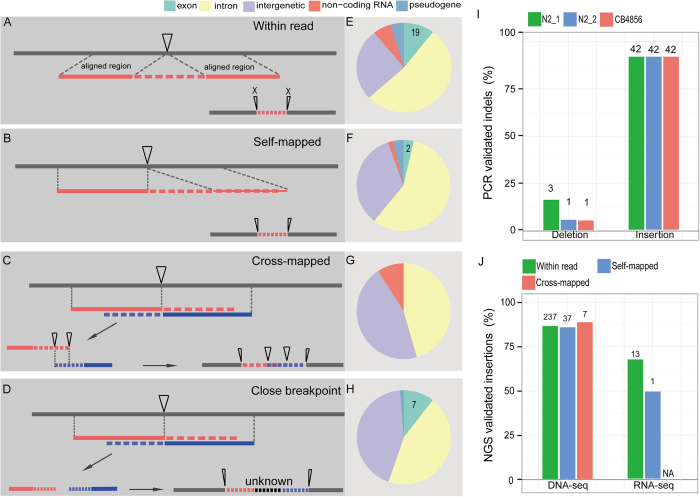
Figure 5. Identification and validation of the long indels (>=9 bp) by PCR and/or NGS data.
(A-D) Shown are the definitions of four possible ways of detecting insertion (insertion position indicated with an inverse triangle). (A) “Within read” type of insertion is defined as an unalignable region within a read against N2 genome while both arms of the read can be successfully aligned with the same reference genome. (B) “Self-mapped” type of insertion is similarly defined as that in (A) except that a second arm can only be aligned with the N2 genome using more permissive alignment parameters (see Methods). (C) “Cross-mapped” type of insertion is defined as an insertion recovered by the overlapping unalignable ends from two independent reads located adjacently in the reference genome. Both unalignable ends that can be partially aligned against N2 genome that are adjacent to each other. (D) “Close breakpoint” type of insertion is similarly defined as that in (C) except that the recovered insertion contains a gap with unknown length due to non-overlapping part between the two unalignable ends. (E-H) Genomic distribution of each type of insertion identified in (A-D). Genomic regions are classified based on their coding potential and differentially colored on the top. Number of insertions located inside coding exon is indicated. (I) Validation results for 51 insertions identified above and 19 deletions identified within long reads by PCR. Shown are the percentages of validated indels that are detected using three different sources of DNA samples as color coded. The numbers of the validated events are indicated above each bar. N2_1 and N2_2 are two N2 C. elegans strain that have been independently maintained over 20 years. CB4856 is the Hawaii isolate of C. elegans. (J) Validation results of three types of insertion as identified in (A, B and C) by NGS data (see Methods). The number of the validated events is indicated above each bar. NA, not applicable.