Figure 4.
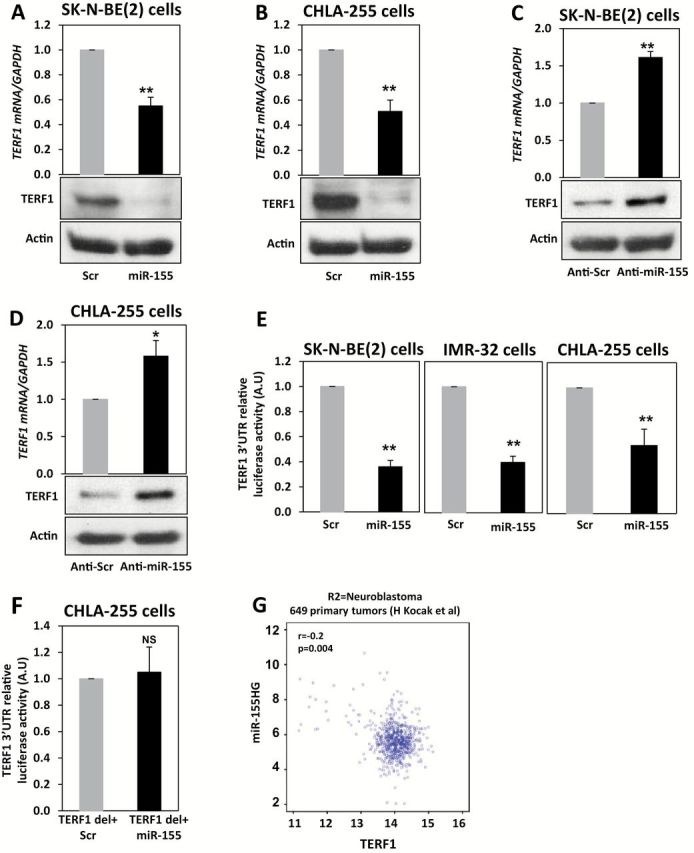
miR-155 and TERF1 regulation in neuroblastoma (NBL). A) Detection of TERF1 mRNA (by quantitative real-time polymerase chain reaction [qRT-PCR]) and protein (by immunoblotting) levels in SK-N-BE(2) cells transfected with a scrambled oligonucleotide (scr group) or with miR-155 (miR-155 group) after 48 hours. B) Same experiment described in (A) but in CHLA-255 NBL cells. C) Detection of TERF1 mRNA (by qRT-PCR) and protein (by immunoblotting) levels in SK-N-BE(2) cells transfected with LNA-anti-miR-155 (anti-miR-155 group) or LNA-anti-scrambled (anti-scr group) oligonucleotide after 48 hours. D) Same experiment described in (C) but in CHLA-255 NBL cells. Relative levels of TERF1 mRNA expression were normalized to GAPDH mRNA whereas β-actin was used as loading control for TERF1 protein. E) Luciferase reporter assay in SK-N-BE(2), IMR-32, and CHLA-255 NBL cells cotransfected with a plasmid expressing the wild-type 3’-UTR of the TERF1 gene downstream of the reporter gene and with miR-155 or a scrambled oligonucleotide and detected after 24 hours. F) Luciferase reporter assay in SK-N-BE(2) cells cotransfected with a plasmid in which the predicted binding site for miR-155 in the 3’-UTR of the TERF1 gene had been deleted (TERF1 del+miR-155) and with miR-155 or a scrambled oligonucleotide and detected after 24 hours from transfection. Data are presented as mean ± SD of experiments conducted in quadruplicate. *P = .009, **P < .001. Student’s t test, two-sided. G) Correlation between miR-155 host gene (miR155HG) and TERF1 mRNA in 649 NBL primary tumors, as detected from a publicly available R2 database. NS = not statistically significant.
