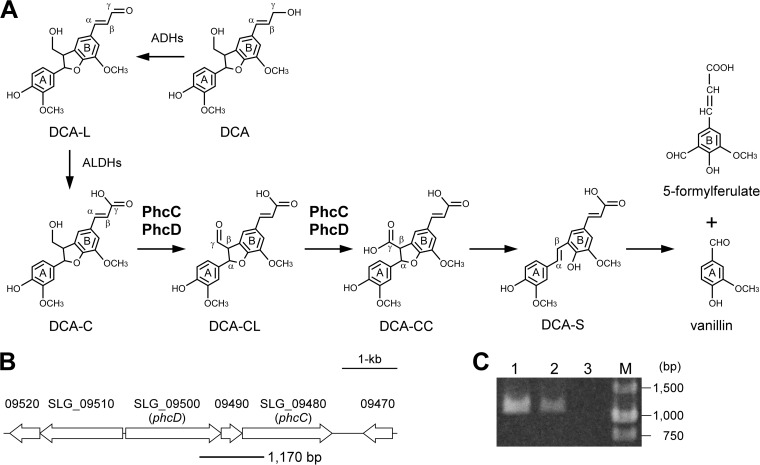
FIG 1.
(A) Proposed catabolic pathway of DCA in Sphingobium sp. strain SYK-6. Enzymes: ADHs, alcohol dehydrogenases; ALDHs, aldehyde dehydrogenases; PhcC and PhcD, DCA-C oxidases. (B) Gene organization of SLG_09480 (phcC) and SLG_09500 (phcD) involved in the oxidation of DCA-C. Arrows indicate the genes from SLG_09470 to SLG_09520. The thick line under the map indicates the location of amplified RT-PCR product shown in panel C. (C) Agarose gel electrophoresis of RT-PCR products (a phcC-phcD intergenic region) amplified with primers shown in Materials and Methods using the following templates: total DNA of SYK-6 (lane 1), reverse transcripts of total RNA of SYK-6 (lane 2), and total RNA of SYK-6 (lane 3). M, molecular size markers.