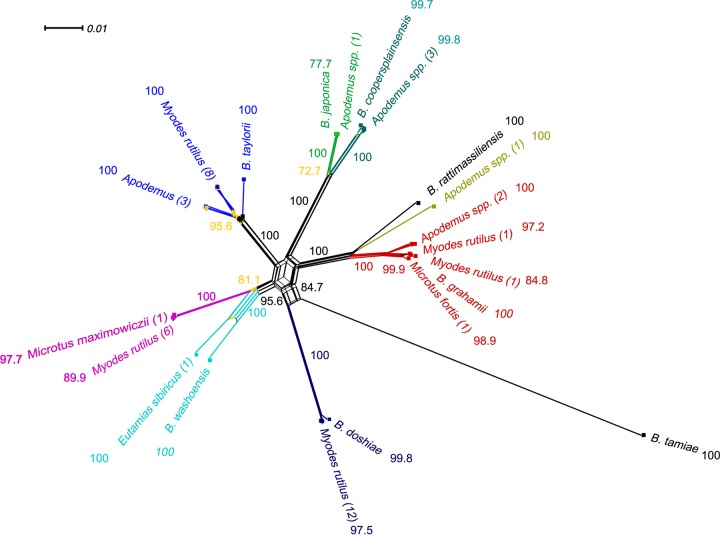
FIG 3.
Neighbor-Net tree of associations of Bartonella isolates obtained from different rodents inhabiting Heixiazi Island, China, and the Bartonella type strains based on concatenated conserved genes (gltA, 16S rRNA gene, ftsZ, and rpoB) representing a total of 3,110 nucleotides using SplitsTree4. The numbers in parentheses represent the number of isolates at each branch. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches; only bootstrap values of >70% are indicated. Different colors refer to individual Bartonella genetic groups. Recombination is illustrated by the reticulation structures in the center among Bartonella strains. Bartonella tamiae was used as an outgroup. The fit parameter values were 98.2% in all cases, indicating that a substantial fraction of the phylogenetic signals could be depicted on the graph. The bar in the upper left corner of the network graph indicates the scale of branches.