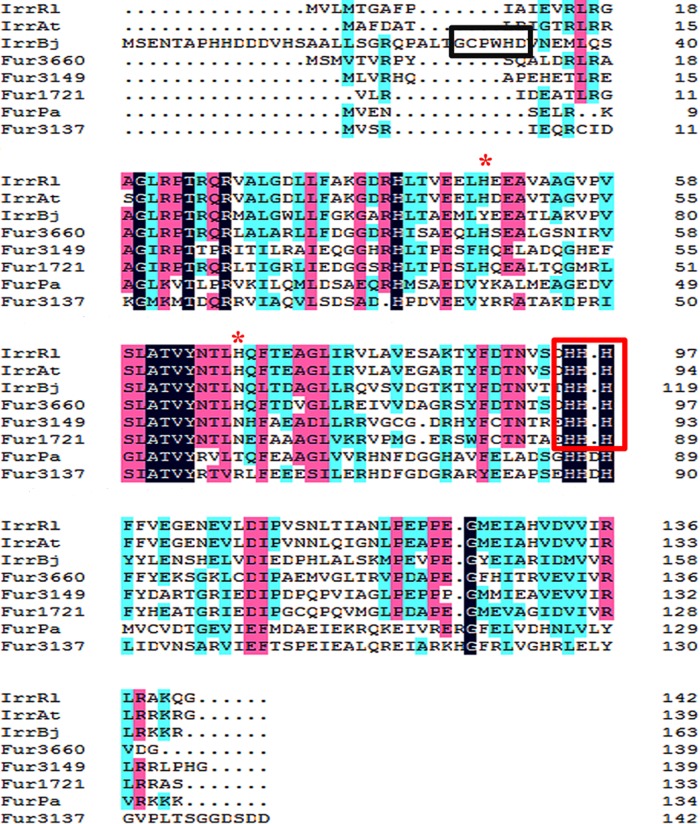
FIG 1.
Primary sequence alignment of Fur and Irr proteins by the ClustalW program. Four Fur homologues were from M. gryphiswaldense MSR-1: MGMSRv2_3137 (Fur; GenBank accession no. YP_008939008), MGMSRv2_1721 (IrrA; GenBank accession no. YP_008937596), MGMSRv2_3149 (IrrB; GenBank accession no. YP_008939020), and MGMSRv2_3660 (IrrC; GenBank accession no. YP_008939529). One related Fur sequence was from P. aeruginosa: Fur-Pa (GenBank accession no. NP_253452). Three related Irr sequences were from B. japonicum (Irr-Bj; accession no. YP_005605653), R. leguminosarum (Irr-Rl; accession no. WP_017962725), and A. tumefaciens (Irr-At; accession no. WP_003493135). Black, pink, or blue shading indicates that eight, six, or fewer than five proteins, respectively, share the same amino acids at a given site. The conserved histidine residue motif (HHH) is boxed in red. The heme regulatory motif is boxed in black. The second heme-binding site is indicated by red asterisks.