Abstract
The conversion of the prion protein (PrPC) into prions plays a key role in transmissible spongiform encephalopathies. Despite the importance for pathogenesis, the mechanism of prion formation has escaped detailed characterization due to the insoluble nature of prions. PrPC interacts with copper through octarepeat and non-octarepeat binding sites. Copper coordination to the non-octarepeat region has garnered interest due to the possibility that this interaction may impact prion conversion. We used X-ray absorption spectroscopy to study copper coordination at pH 5.5 and 7.0 in human PrPC constructs, either wild-type (WT) or carrying pathological mutations. We show that mutations and pH cause modifications of copper coordination in the non-octarepeat region. In the WT at pH 5.5, copper is anchored to His96 and His111, while at pH 7 it is coordinated by His111. Pathological point mutations alter the copper coordination at acidic conditions where the metal is anchored to His111. By using in vitro approaches, cell-based and computational techniques, we propose a model whereby PrPC coordinating copper with one His in the non-octarepeat region converts to prions at acidic condition. Thus, the non-octarepeat region may act as the long-sought-after prion switch, critical for disease onset and propagation.
The pathological form of the cellular prion protein (PrPC) plays a central role in a variety of human and animal neurodegenerative disorders, known collectively as prion diseases or transmissible spongiform encephalopathies (TSE). These disorders arise from misfolding of the mainly α-helical folded PrPC to a β-sheet enriched, protease-resistant and infectious isoform, termed PrPSc or prion. TSE uniquely manifest as sporadic, genetic and iatrogenic forms, and they include most notably Creutzfeldt-Jakob disease in humans, bovine spongiform encephalopathy in cattle, scrapie in sheep and goat, and chronic wasting diseases in cervids1.
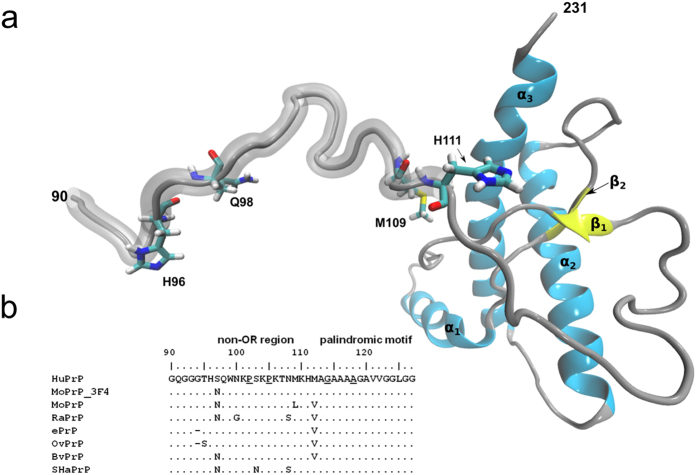
Structural information from NMR spectroscopy and X-ray crystallography studies shows that the C-terminal PrPC domain (residues 128 to 231, hereafter in human PrPC numbering) adopts a predominantly α-helical conformation with small β-sheet content2,3 (Fig. 1a). The N-terminal half of the protein (residues 23 to 127) does not adopt any identifiable folding structure in solution4. The latter features four histidine (His) residues within the highly conserved octapeptide repeats (OR) (residues 60–91). This region is able to coordinate up to four copper ions with distinct coordination geometries and high affinity5. Two additional His residues able to bind copper are found in the 92–111 region, including a charged segment (residues 92–111) denoted as non-OR region6 adjacent to a hydrophobic segment (residues 112–127) thought of playing a role during prion conversion7. The amino acidic sequence spanning residues 90 to 127 shows high degree of conservation among different mammalian species, with local amino acidic substitutions clustered in the non-OR region (Fig. 1b).
Figure 1. The non-OR region in the truncated HuPrP.
In (a) cartoon representation of the HuPrP(90–231) with highlighted the segment 90–111 including the apo form of the non-OR copper binding site. Residues H96, Q98, M109 and H111 contribute to coordinate one Cu(II) or Cu(I) ion. In (b), alignment of residues 90–127 including the non-OR region and the palindromic motif of different mammalian prion proteins (PrP): HuPrP (human, Homo sapiens, NCBI accession code AAA60182), MoPrP (mouse, Mus musculus, AAA39997) and MoPrP_3F4 (carrying the 3F4 tag), RaPrP (rabbit, Oryctolagus cuniculus, AAD01554.1), ePrP (elk, Cervus elaphus nelsoni, AAB94788), OvPrP (sheep, Ovis aries, AFM91142.1), BvPrP (bank vole, Myodes glareolus, AAL57231) and SHaPrP (Syrian hamster, Mesocricetus auratus, AAA37091). In the HuPrP primary sequence the residues involved in GSS-linked mutations are underlined (P102L, P105L, G114V and A117V).
Unlike PrPC, the PrPSc structure features a significant –if not all- content of β-sheet secondary structure motives8. These unique structural features of PrPSc are responsible for its physicochemical properties, including insolubility in non-ionic detergents, protease-K (PK) resistance and aggregation propensity1. The insoluble and heterogeneous nature of PrPSc renders its structural characterization extremely difficult, and to date several prion structural models have been proposed9.
Yet, normal PrPC function(s), as well as the structural and molecular mechanisms leading to PrPSc conversion and transmission, remain largely unknown10. PrPC is tethered to the extracellular cell surface via a modified lipid anchor linked to the C-terminus, and it displays highest expression levels at the presynaptic membrane of neurons11. Mounting evidence suggests a central role for PrPC in neuroprotection. Proposed PrPC functions range from neuronal growth and differentiation12, synaptic plasticity13, cell signaling14, to N-methyl-D-aspartate (NMDA) receptors modulation15,16,17 and brain metal homeostasis18. The latter two functions are attributed to the PrPC ability to bind copper and -to a lesser extent- other divalent cations such as zinc and manganese19,20. At the synapse, copper is released during depolarization via synaptic vesicles, reaching transient concentrations up to 250 μM in the synaptic cleft, thus suggesting that copper release may be an integral component of neurotransmission21. Copper binding to PrPC promotes its endocytosis via clathrin-dependent pathway, arguing that this endocytic mechanism may serve to reuptake copper within the presynaptic terminal after depolarization22. More recent results confirmed that PrPC and copper cooperatively exert neuroprotection limiting intracellular calcium accumulation through a copper-mediated S-nitrosylation of the NMDA receptor15.
Although copper binding seems to be involved in many physiological PrPC processes, the interplay between copper and PrPSc conversion poses intriguing issues. While the OR region does not appear to be essential for prion infectivity23, the non-OR copper-binding site may be relevant because of its location adjacent to a region –the palindromic motif sequence AGAAAAGA- involved in structural changes during the early stage of prion conversion24. This regional proximity raised the question whether copper bound to the non-OR site may have an impact on prion conversion. Indeed, the interaction of copper with a peptide including both the non-OR and the palindromic motif has been shown to induce β-sheet formation and aggregation of this segment25,26. Further in vivo findings have supported the idea that the region encompassing residues 90–125 is involved in prion generation. In addition, this region seems to have a crucial role in preserving physiological PrPC function(s). While the deletion of the OR in transgenic (Tg) mice is not toxic, the ablation of segments including the non-OR and the hydrophobic region results in diseased mouse phenotypes with cerebral disorders27,28. Interestingly, naturally occurring point mutations clustered in the non-OR region and in the palindromic motif are responsible for Gerstmann–Sträussler–Scheinker (GSS) syndrome, characterized by PrPSc amyloidal plaque deposits in the brain (Fig. 1b). These observations corroborate the idea of the non-OR as pivotal partner in both PrPC function and conversion to prion.
Here, by means of extended X-ray absorption fine structure (EXAFS) spectroscopy, cell-biology approaches and molecular dynamic (MD) simulations, we have investigated how copper coordination in the non-OR region may influence prion conversion. EXAFS spectroscopy is a powerful technique for obtaining structural and chemical information about metal binding to a protein of interest, providing accurate bond length measurements within 5 Å or less. Several groups have analyzed by EXAFS copper coordination bound to recombinant PrP in the OR29 and non-OR regions30,31. In a previous EXAFS study we found that at pH 5.5 Cu(II) and Cu(I) bound to wild-type (WT) are specifically coordinated by the Nδ-atom in the imidazole ring of two His residues (H96 and H111), by deprotonated amide nitrogens from adjacent residues (e.g. Q98), as well as by sulfur-mediated bounds from either methionine (Met) 109 or M11130. This coordination geometry is completely altered in the HuPrP(90–231) carrying the Q212P substitution, a mutation linked to GSS syndrome, suggesting that this mutant may have structural consequences both at the C-terminal globular domain32 and at the non-OR region. In order to gain insights into the role of mutations on the non-OR region, we investigated the copper coordination in the WT HuPrP(90–231) and in different mutants at both pH 7.0 and pH 5.5 – the latter mimicking the acidic endosomal compartments33. As model systems for the mutants we used Q212P, P102L –the prototypical GSS mutation34 - and H96Y –an artificial mutation devoid of one His residue involved in copper binding. EXAFS data clearly highlighted modifications of the non-OR copper-binding site induced by these mutations. To understand the physiological implications of our EXAFS data, we performed in vitro and cell-based approaches. We used neuronal cell models expressing 3F4-tagged murine (Mo) PrPC (MoPrP) (Fig. 1b) in which the N-terminal His residues within the OR and non-OR regions were substituted by tyrosine (Tyr) and we evaluated the effect of single His to Tyr substitutions in the context of prion conversion. Intriguingly, only H96Y mutation largely promoted prion conversion, PrP accumulation in the endosomal compartments, and generation of bona fide infectious prion material (e.g. displaying partial PK-resistance and the ability to perform seeded aggregation in vitro and in cell). Finally, MD simulations were used to hypothesize possible structural consequences on the HuPrP structure caused by an altered copper coordination in the non-OR region. In our simulations we predicted structural facets of WT HuPrP(90–231) coordinating Cu(II) via either H111 or both H96 and H111. Comparison of trajectories showed that removal of H96 ligand from the Cu(II) coordination results in β-sheet enrichment in the segment spanning the non-OR region and palindromic motif.
Our study highlights the importance of the non-OR region for prion conversion and proposes a model in which PrPC coordinating copper with one His residue may render PrPC more prone to prion conversion at acidic pH condition.
Materials and Methods
Plasmids construction, protein expression and purification
The H96Y, P102L and Q212P mutations were inserted in pET-11a encoding for HuPrP(90–231) as previously described30. The pET-11a plasmid (Novagen) encoding for the full-length MoPrP -residues 23–231, here denoted as MoPrP(23–231)- was kindly provided by Dr. J.R. Requena (University of Santiago de Compostela, Santiago de Compostela, Spain). The H96Y mutation was inserted into the full-length MoPrP DNA by mutagenesis kit (Stratagene). The open reading frame encoding for the pre-pro MoPrP from residue 1 to 254 was amplified by PCR from genomic murine DNA and cloned by restriction-free method in pcDNA3.1(-) vector (Invitrogen). The 3F4 epitope tag and the single point mutations (H61Y, H69Y, H77Y, H85Y and H96Y in human numbering single amino acid code) were inserted by mutagenesis into the pcDNA3.1::MoPrP(1–254). The WT HuPrP(90–231), HuPrP(90–231, H96Y), HuPrP(90–231, P102L), HuPrP(90–231, Q212P) and the WT MoPrP(23–231) were expressed, purified and in vitro refolded according to our previous protocols30,32.
XAS spectra data collection
Samples with 1:1 Cu(II):WT and mutants HuPrP(90–231) ratio were prepared in 25 mM NaOAc pH 5.5 and 25 mM MOPS buffer pH 7.0. The Cu(I):WT and mutants HuPrP(90–231) complexes were generated reducing Cu(II) with 40 mM ascorbate as previously described30. X-ray absorption spectra were recorded at the European Synchrotron Radiation Facility (ESRF) on beam line BM30B FAME35, under ring conditions of 6.0 GeV and 180 mA. The spectra were collected at the Cu K-edge in fluorescence mode using a solid state 30-element Ge detector, with sample orientation at 45° to incident beam. The X-ray photon beam was vertically focused by a Ni−Pt mirror, and dynamically sagittally focused in the horizontal size. The monochromator was equipped with a Si(111) double crystal, in which the second crystal was elastically bent to a cylindrical cross section. The energy resolution at the Cu K-edge is 0.5 eV. The spectra were calibrated by assigning the first inflection point of the Cu foil spectrum to 8981. All the spectra were collected at 10 K. For the Cu(II) samples photo reduction was observed and thus the beam was moved to different spots of the sample at each scan. During collection, data were continuously monitored in order to insure sample homogenity across the multiple spots collected from different sample-holder’s cells. Complete data sets were collected for samples 1 to 14 (Supplementary Table 1). For each sample, 12 spectra were recorded with a 7 s/point collection statistic and averaged. The collection time was 25 min for each spectrum.
EXAFS data analysis
EXAFS data analysis was performed using the GNXAS code, which is based on the decomposition of the EXAFS signal (defined as the oscillation with respect to the atomic background cross-section normalized to the corresponding K-edge channel cross-section) into a summation over n-body distribution functions γ(n), calculated by means of the multiple-scattering (MS) theory36. The analysis of the EXAFS spectra was carried out starting from the coordination model previously determined30. In particular, Cu(II) was found to be coordinated to H96, H111, and two additional low Z ligands (oxygen or nitrogen donors) in the inner shell and to a sulphur-donating ligand, which was assigned to M109. The final fit included also one close oxygen-donating ligand that was assumed to derive from solvent, although it could also be derived from a protein ligand such as a Q98. Based on this model, theoretical EXAFS spectra were calculated to include contributions from first shell two-body signals, and three-atom configurations associated with the His rings. The model χ(k) signal has been then refined against the experimental data by using a least-squares minimization procedure in which structural and nonstructural parameters are allowed to float. The structural parameters are bond distance (R) and bond variance (σ2R) for a two-body signal, the two shorter bond distances, the intervening angle (θ), and the six covariance matrix elements for a three-body signal. During the minimization procedures, the magnitudes of the Debye–Waller terms for the imidazole rings were assumed to increase with distance, and imidazole ring atoms at similar distances from the copper ion were assigned the same value. The EXAFS spectra were analyzed trying out different possible models including either one or two His ligands and a sulfur coordinating atom. In all cases two additional nonstructural parameters were minimized, namely, E0 (core ionization threshold) and S02 (many body amplitude reduction factor). The quality of the fits was determined by the goodness-of-fit parameters37, Ri, and by careful inspection of the fit of the EXAFS data, Fourier trasform, and individual EXAFS γ(n) signals. To establish error limits on the structural parameters, a number of selected parameters from the fit results were statistically analyzed using two-dimensional contour plots. This analysis examines correlations among fitting parameters and evaluates statistical errors in the determination of the copper coordination structure, as previously described36. Briefly, parameters with highest correlation dominate in the error estimate. The results of the EXAFS fits for all the systems are given in Supplementary Table 1 and the best-fit curves are shown in Supplementary Fig. 1 for Cu(II) and Cu(I) HuPrP(90–231) at pH 7.0, as an example. The EXAFS spectra were analyzed in the k range between 2.4–12.6 Å−1 for Cu(II) and 2.5–13.0 Å−1 for Cu(I). In all cases S02 was found equal to 0.9, while E0 was found 3 and 7 eV above the first inflection point for the Cu(II) and, Cu(I) spectra, respectively.
Neuroblastoma cell cultures
N2a and ScN2a cells were cultured in Opti-MEM (GIBCO) media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin, and incubated at 37 °C, 5% CO2. Transient transfections were performed using X-treme gene DNA transfection kit (Roche Biochemicals) according to the manufacturer guidelines. Seventy-two hours post-transfection, the cells were collected for further analysis.
Biochemical assays on PrPSc and PrPC
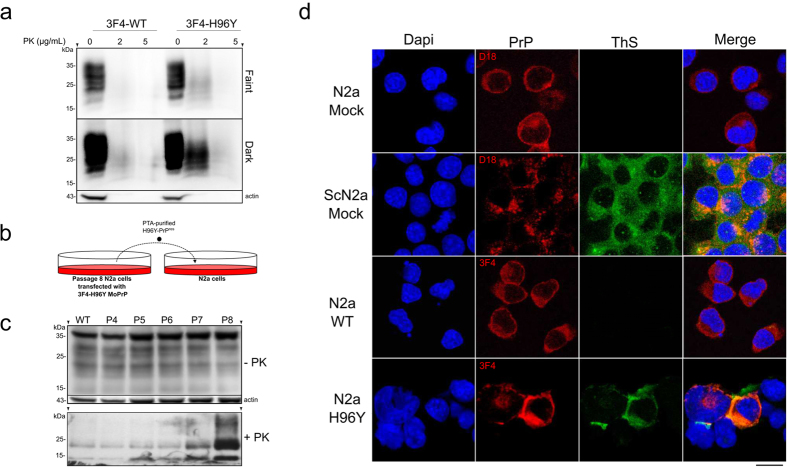
Cell lysates were harvested in cold lysis buffer (10 mM TrisHCl pH 8.0, 150 mM NaCl, 0.5% Nonidet P-40 substitute, 0.5% sodium deoxycholate), quantified by BCA protein assay kit (Pierce) and stored at −20 °C until use. For protease-K (PK) digestion assay, quantified protein lysates were treated with PK (Roche) at 37 °C. ScN2a cell lysates were digested with 20 μg/mL of PK for 1 hour, while cell lysates from N2a cells transfected with either WT or H96Y 3F4-MoPrP constructs were digested with 2 and 5 μg/mL of PK for 30 minutes. PK digestions were stopped by adding 2 mM phenylmethyl-sulphonyl fluoride. Subsequently, the samples were ultracentrifuged at 100,000 g for 1 h at 4 °C (Optima TL, Beckman Coulter, Inc.). The pellets were resuspended in sample buffer. The glycan modifications on PrPC were assessed using Endo-H and PNGase-F enzymes (New England Biolabs) according to the manufacturer instructions. Samples were loaded onto a 10% SDS-PAGE gel and immunoblotted on Immobilion PVDF (Millipore) membranes. Membranes were blocked with 5% (w/v) non-fat milk protein in TBS-T (0.05% Tween), incubated with 1:1000 anti-PrP antibody 3F4 (Covance), and developed by enhanced chemiluminescence (GE Healthcare). Band intensity was acquired using the UVI Soft software (UVITEC, Cambridge). Total PrPC expression levels in N2a and ScN2a cell lysates were normalized on β-actin value using 1:10,000 anti-β-actin Peroxidase (Sigma-Aldrich). The PrPSc PK-resistance levels in all the mutants were derived using as reference the PK-resistant band intensity of the ScN2a cells transfected with WT 3F4-MoPrP. To evaluate the role of copper in prion conversion, ScN2a cells were transiently transfected with pcDNA3.1::3F4-MoPrP(1–254) WT and pcDNA3.1::3F4-MoPrP(1–254) H96Y plasmids, treated for 48 hours with increasing concentration (10, 20, 30 and 40 μM) of cuprizone (CPZ, Sigma Aldrich) and immediately collected for lyses, PK digestion and immunoblot as described above.
Fluorescence imaging
Cells were grown on poly-L-lysine-coated coverslips for 24 hours before fixation in 4% paraformaldehyde and washed with PBS prior to blocking in 1% FBS, 0.3% Triton X-100. Cells were incubated at 4 °C for 12 hours in blocking buffer with anti-PrP primary antibodies, i.e. 3F4 and D18 (InPro Biotechnology) monoclonal antibodies. The following day, cells were incubated for 1 hour with secondary antibodies conjugated with AlexaFluor. For PrPC cell surface detection, cells were incubated at 4 °C for 15 min and probed with 3F4 antibody. Cells were permeabilized with 0.2% Triton-X and stained with AlexaFlour-488 secondary antibody. To detect Thioflavin-S (ThS)-positive aggregates, transfected and non-transfected cells were fixed with 4% PFA/4% sucrose/1% Triton X-100 in PBS. For organelle markers, we used anti-Calnexin (ER marker), anti-EEA1 (early endosome marker), anti-Tfn (recycling endosome marker), anti-M6PR (late endosomes marker) and anti-LAMP2 (lysosomes marker) purchased from Abcam. Nuclei were stained with DAPI dye (VECTOR Laboratories). Images were acquired with a DMIR2 confocal microscope equipped with Leica Confocal Software (Leica).
Monitoring the kinetics of in vitro fibril formation
To monitor the formation of ThT-positive fibrils, we used 100 μg/mL of WT full-length MoPrP stored in acetate buffer (25 mM NaOAc, 6 M GdnHCl, pH 5.5) or Tris buffer (25 mM Tris-HCl, 6 M GdnHCl, pH 7.0). To induce protein polymerization in the amyloid seeding assay (ASA) we added a preformed PrPSc seed to the reaction purified from ScN2a transiently expressing either 3F4-tagged WT MoPrP or 3F4-tagged H96Y MoPrP according to our previous protocols. Data were analyzed and figures were produced using Origin 8.6 software.
Prion formation in N2a cells
N2a cells were transiently transfected with either 3F4-tagged WT or H96Y MoPrP and regularly passaged every 7 days up to passage (P) 8. Subsequently, the protein extracts were analyzed by PK digestion to monitor the presence of PK-resistant PrPSc levels through passages (see above). Prion formation was assessed by cell seeding experiments. Phosphotungstic acid (PTA)-extracted PrPres seeds -we denoted as PrPres the material generated by N2a cells transfected with 3F4-tagged H96Y MoPrP- were subjected to N2a cells and regularly passaged every 7 days up to P8. The detection of newly generated PrPres was assessed by PK digestion as previously described, and the PrPres seeds isolation using PTA was performed according to our previous protocols.
Building of MD system and simulation protocol
Two simulations of WT HuPrP(90–231) in complex with Cu(II) were performed in order to resemble experimental conditions either at pH 5.5 or pH 7.0. Based on experimental results, in the simulation resembling pH 5.5 (hereafter termed 2His) both H96 and H111 were bound to Cu(II), while at pH 7.0 (hereafter termed 1His) only H111 was bound to the metal. Starting coordinates of HuPrP(90–231) were obtained from the 1QM1 PDB structure4. Missing hydrogen atoms were added by MolProbity38 and protonation states counter-checked with the H++ code39. To model the binding of Cu(II) in the active site a set of harmonic springs was used to enforce copper-ligand distances40. Bond lengths of the springs were based on EXAFS data and set to 1.98 Å for the Nδ imidazole atoms in H96 and H111, to 1.98 Å for the amide oxygen in Q98 and to 3.25 Å for the S atom in M109. The systems were modeled using the AMBER99SB-ILDN* all-atom force field41 implemented in the GROMACS MD package version 4.6.542. The protein was immersed in a rhombic dodecahedron box with a minimum distance of 2.5 nm from the box edges; a long distance from the box edges was used to take into proper account the flexibility of HuPrP(90–231). The resulting system was composed of 2192 protein atoms, 54873 TIP3P43 water molecules, 1 Cl− ion for a total of 166812 atoms. Electrostatic interactions were accounted by means of the Particle Mesh Ewald method (PME) using a cutoff of 1.5 nm for the real space and Van der Waals interactions44. The LINCS algorithm was used to constrain bond lengths and angles45, with the exception of Cu(II)-protein bonds. Relaxation of solvent molecules and Cl− ion was initially performed keeping solute atoms restrained to their initial positions with a force constant of 1000 kJ/(mol • nm2), for 3.0 ns in a NPT ensemble and using an integration time step of 1.0 fs. Then, the system was simulated for 5 ns while using a force constant of 1000 kJ/(mol • nm2) for bonds involving Cu(II) to force the active site residues in the desired position. At this point H111 was freed to create the 1His system and after this step two separate simulations were carried out. The two systems were carried again to 0 K, the force constants in the active site changed to 100 kJ/(mol • nm2) and then heated again to 298.15 K. The two systems were then simulated for 150 ns in a NVT ensemble with a time step of 2.0 fs and the neighbor list was updated every 10 steps. Temperature was kept constant using the velocity rescale method with a coupling constant of 0.1 ps during sampling46. Structures were clustered using backbone atoms and the GROMOS method47. A total of 15000 frames from each simulation were selected with constant pace of 10 ps and a Root Mean Square Deviation (RMSD) threshold of 0.30 nm; only the positions of Cα carbon atoms were used for clustering. Two residues were considered to be bound by a hydrogen (H) bond in a given frame if the H-acceptor distance was below 0.375 nm and the acceptor – donor– H angle was below 40.0°. Two residues are connected by a salt bridge if the nitrogen-oxygen distance is below 0.4 nm in a given frame. All analyses were carried out with standard tools present in the GROMACS MD package v. 4.6.5 or with in-house written codes, except for secondary structure assignment which was performed with DSSP48. Figures were produced with Chimera 1.10 software.
Results
Copper coordination in the non-OR region of WT HuPrP
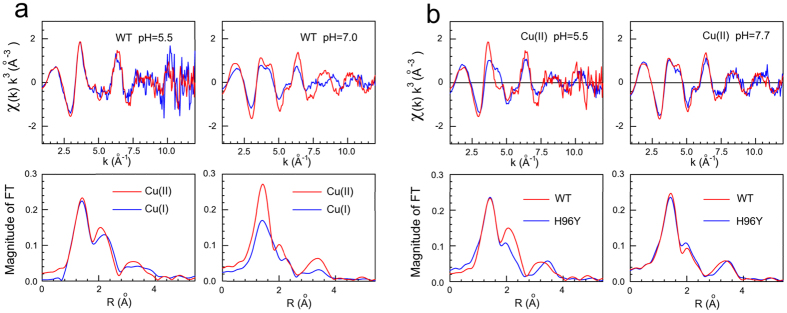
A previous XAS investigation was used to determine the atomic structure of non-OR copper-binding site in the WT HuPrP(90–231)30; in this work XAS experiments at the Cu K-edge were carried out at pH 5.5 and the structure of the binding site of both Cu(II) and Cu(I) was found to be identical. Quantitative analysis of the EXAFS spectra indicated that in both oxidation states, copper ion is coordinated by two His residues (H96 and H111 with Cu-N distance of 1.98(2) Å), by two low Z ligands (either oxygen or nitrogen atoms at 1.98 (1.99) Å) and by one sulphur scatterer at longer distance (3.25 Å)30. To highlight the effect of pH on the local coordination of the copper ion, XAS spectra of both Cu(II) and Cu(I) HuPrP(90–231) complexes were collected at pH 7.0 (Fig. 2a). While at pH 5.5 the EXAFS spectra are almost identical for both copper oxidation states, at pH 7.0 EXAFS signals show markedly different features over the full k-range. The quantitative analysis of the EXAFS data indicates that at pH 7.0 only a single His coordinates the metal ion in both oxidation states, thus suggesting that one of the two His residues (H96 or H111) moves away from the metal. EXAFS data concerning Cu(II)-HuPrP(90–231) at pH 7.0 could be modeled as a four coordinate copper center with one His at 1.99(2) Å, and three N/O scatterers at 1.99(4) Å with a more distant sulphur scatterers at 3.47(4) Å. As far as the Cu(I)-HuPrP(90–231) protein is concerned, the EXAFS analysis revealed a three-fold coordination with one His at 1.98(2) Å, one N/O scatterers at 2.00(2) Å and one sulphur scatterer at 2.27(4) Å (Supplementary Fig. 1). In this case one Met residue enters the Cu(I) first coordination shell, similarly to what previously found at acidic pH value30.
Figure 2. Copper coordination in the non-OR region of WT and H96Y HuPrP.
k3-weighted EXAFS spectra and Fourier transforms of the experimental data of Cu(II) and Cu(I) bound to WT HuPrP(90–231) at pH 5.5 and 7.0 (a), and of Cu(II) bound to WT HuPrP(90–231) and H96Y at pH 5.5 and 7.0 (b).
Copper coordination in H96Y, P102L and Q212P HuPrP mutants
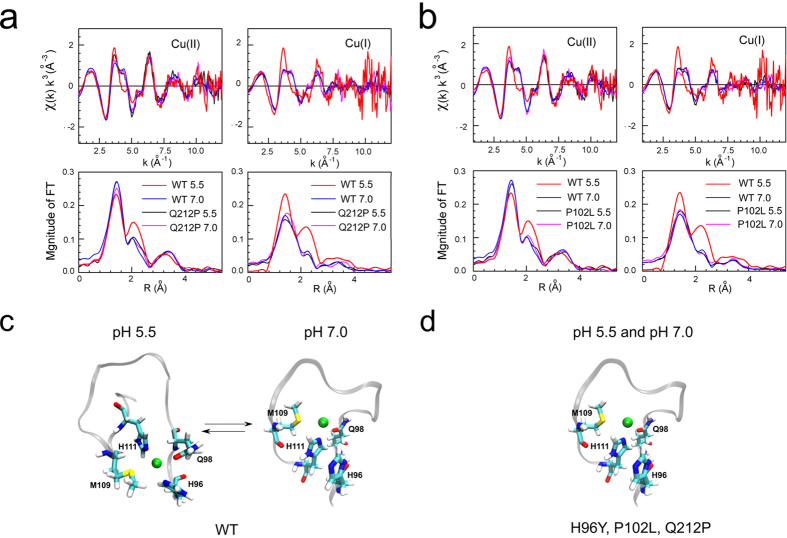
The Cu(II) local coordination structure in the H96Y mutant was investigated both at pH 5.5 and 7.0 values. At pH 5.5, the Cu K-edge EXAFS data for H96Y, compared to the WT protein, exhibited a clear modification in the coordination environment. This variation is explained by the existence of a single His in the non-OR region of the H96Y mutant, hampering the coordination of the Cu(II) ion with two His residues (Fig. 2b). Conversely, at pH 7.0 the EXAFS spectra of the H96Y mutant and WT proteins are almost identical, thus suggesting that H111 is involved in the copper binding site in both cases. In addition, the EXAFS data showed that the H96Y mutant maintains the same coordination environment around the Cu(II) ion when increasing pH from 5.5 to 7.0. The analysis of the EXAFS data revealed the presence of a four coordinate copper center almost identical to that of the WT protein at pH 7.0, with the H111 residue at 2.00 (2) Å from the ion (Supplementary Table 1). The structures of Cu(II) and Cu(I) binding sites were also investigated in Q212P and P102L HuPrP mutants at both pH 5.5 and 7.0 values. The EXAFS data of the mutants share the same coordination pattern observed for the WT at pH 7.0, where the copper ion is coordinated by a single His, namely H111 (Fig. 3a,b and Supplementary Table 1). In conclusion, while in the WT the copper ion -at both oxidative states- changes coordination losing the contact with H96 at pH 7.0 (Fig. 3c), in the mutants copper is bound only to H111 independently of the pH (Fig. 3d).
Figure 3. Comparison between copper K-edge XAS experimental data of pathological Q212P, P102L and WT HuPrP.
k3-weighted EXAFS spectra and Fourier transforms of Cu(II) and Cu(I) bound to WT HuPrP(90–231) and Q212P at pH 5.5 and 7.0 (a) and of Cu(II) and Cu(I) bound to WT HuPrP(90–231) and P102L at pH 5.5 and 7.0 (b). Schematic representations of copper binding sites in the WT HuPrP(90–231) (c) and in the mutants (d) at both pH 5.5 and 7.0. Green spheres identify a single cupper ion at both oxidative states.
The H96Y mutation promotes prion conversion in neuroblastoma cells
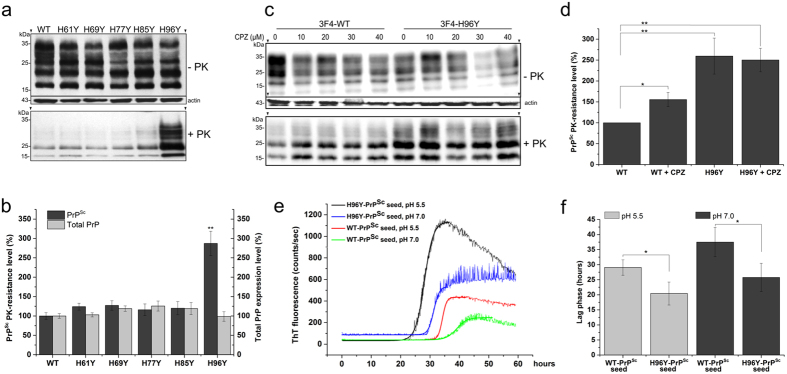
The observed structural differences in the copper coordination among WT and pathological mutants at pH 5.5 may have relevant physiological implications since this alteration in the copper binding site might trigger PrPC to PrPSc conversion. Hence, the non-OR region could be an important “hot spot” for prion conversion. We first investigated the effect on prion replication of single His residues along the entire N-terminal PrPC domain. ScN2a cells were transiently transfected with 3F4-tagged WT and mutant MoPrP constructs in which each individual His located inside the OR and non-OR copper binding sites were substituted by Tyr, thus abolishing the physiological copper binding. The introduction of the 3F4-epitope tag into these constructs makes it possible to discriminate between transfected and endogenous PrPC. His to Tyr substitutions in MoPrP did not affect the total PrP expression levels (Fig. 4a,b and Supplementary Fig. 2a). The enhanced resistance to protease digestion is a primary feature to discriminate between PrPSc and PrPC in cells chronically infected by prions. The PK digestion profiles showed remarkably different PrPSc levels among the mutants. While other mutants in the OR region displayed negligible PK-resistant PrPSc levels similar to the WT, the H96Y mutant yielded a significantly higher PK-resistant PrPSc signal, providing a first evidence for the role of H96Y mutation in prion conversion (Fig. 4a,b).
Figure 4. The non-OR H96Y mutation promotes prion conversion in ScN2a cells.
(a) Fifty μg of undigested lysates from ScN2a cells expressing 3F4-tagged WT and mutated MoPrPs was applied to each lane. Five hundred μg of cell lysate was digested with PK (20 μg/mL) at 37 °C for 1 hour. MoPrPs were detected by anti-PrP 3F4 antibody. β-actin was used as internal loading control. Arrows (▾) indicate positions on the gels where the blots have been cropped. Full-length blots are presented in the Supplementary Figure S4a–b. (b) Quantitative analysis of total PrP expression and PrPSc PK-resistance levels in transfected constructs (n = 4, **P < 0.005, by two-tailed t test). (c) Copper chelation promoted increased PrPSc formation in 3F4-WT transfected ScN2a cells. CPZ, cuprizone. Full-length blots are presented in the Supplementary Figure S4c–e. (d) Quantitative analysis of PrPSc PK-resistance levels in 3F4-WT and 3F4-H96Y MoPrP transfected ScN2a cells treated with 10 μM CPZ (n = 3, **P < 0.005 and *P < 0.05). (e) ASA showing the kinetics of MoPrP fibrillization in the presence WT and H96Y-PrPSc seeds at pH 5.5 and 7.0 and (f) the corresponding mean value of the lag phases in hours (n = 4, *P < 0.05).
Because H96 binds copper, it is likely that the removal of this copper ligand might render PrPC more prone to the conversion in ScN2a cells. Consequently, the absence of copper from the non-OR region could promote this pathological process. To verify this hypothesis, we measured the PrPSc PK-resistance levels in ScN2a cells transfected either with WT or H96Y MoPrP and treated for 48 hours with cuprizone (CPZ), a well known selective Cu(II) chelator that does not affect cell viability and cannot cross plasma membranes49. CPZ treatments on WT ScN2a cells promoted a significantly increase of PrPSc-PK resistance levels upon 10 to 40 μM CPZ additions, suggesting that PrPC in the apo form is more susceptible to PrPSc conversion (Fig. 4c,d and Supplementary Fig. 2b). The PrPSc levels remained a plateau among control and CPZ-treated H96Y ScN2a cells (Fig. 4c,d) but always higher than PrPSc level in the WT cells as also previously presented (Fig. 4a,b). These data are consistent with the hypothesized mechanism whereby H96Y mutant is per se sufficient to generate high amount of PrPSc molecules. Copper appeared as a pivotal modulator of this process since its absence from PrPC side seems to promote prion conversion.
Biochemical properties of H96Y mutant
We then investigated whether purified PrPSc from H96Y ScN2a cells shares biochemical properties typical of natural prions, including features such as templating the β-sheet conversion of new PrP molecules, partially PK-resistance, positivity to ThT and ThS staining, cell-to cell transmissibility and intracellular accumulation. We therefore evaluated these properties in vitro and in cell-based experiments.
By means of amyloid seeding assay, PTA-isolated PrPSc seeds from ScN2a cells expressing H96Y mutant were used to promote the conversion of recombinant full-length WT MoPrP. Differently from the WT-PrPSc seed, we found that the addition of the H96Y-PrPSc seed significantly promoted MoPrP fibrillization reactions at both pH 5.5 and 7.0 values, resulting in ThT positive kinetics with shorter lag-phases than the controls (Fig. 4e,f). To test the hypothesis that H96Y mutant causes prion formation in non-prion infected cells, N2a cells were transfected with 3F4-WT or H96Y mutant MoPrP and regularly passaged. We found immunoreactive mildly PK-resistant PrP bands –denoted as PrPres or bona fide PrPSc in the absence of in vivo assays- starting from passage (P) 4 to P8 (Fig. 5a and Supplementary Fig. 2c). The isolated H96Y-PrPres material observed in P8 was then used as infectious seed in new N2a cells regularly passaged up to P8 (Fig. 5b). Interestingly, we observed an increment in PrPres levels over passages, thus indicating that H96Y-PrPres seeds induced de novo conversion of endogenous PrPC to PK-resistant PrP material (Fig. 5c). Subsequently, we evaluated whether N2a cells transfected with H96Y mutant displayed tinctorial features reminiscent of PrPSc. By using Thioflavin-S (ThS), a specific dye for staining in cells the protein aggregates enriched in amyloid motifs, we found ThS-positive cytoplasmic H96Y mutant deposits similarly to ScN2a (Fig. 5d).
Figure 5. Biochemical properties and cellular localization of H96Y mutant in N2a cells.
(a) The H96Y mutant displays mild PK-resistance when expressed in N2a cells regularly passaged every 7 days up to passage (P) 8. Cell lysates were treated with 2 or 5 μg/mL of PK. Two exposures of the same blot are shown (Faint: 30 sec exposure; Dark: 6 min exposure). PrPs were detected by anti-PrP 3F4 antibody. β-actin is used as internal control. Arrows (▾) indicate positions on the gels where the blots have been cropped. Full-length blots are presented in the Supplementary Figure S5a–b. (b) Schematic representation of the seeding experiment in N2a cells. (c) PTA-extracted PrPSc from N2a cells transfected with 3F4-H96Y MoPrP were inoculated into N2a cells and regularly passaged every 7 days up to P8. The PrPres detection was assessed by PK digestion (5 μg/mL) through passages. Full-length blots are presented in the Supplementary Figure S5c–d. (d) ThS-positive H96Y MoPrP aggregates detected in N2a cells. The cells were stained for PrP expression (red) and ThS (green). Untransfected N2a and ScN2a cells (mock) were used as controls. Scale bar: 12 μm.
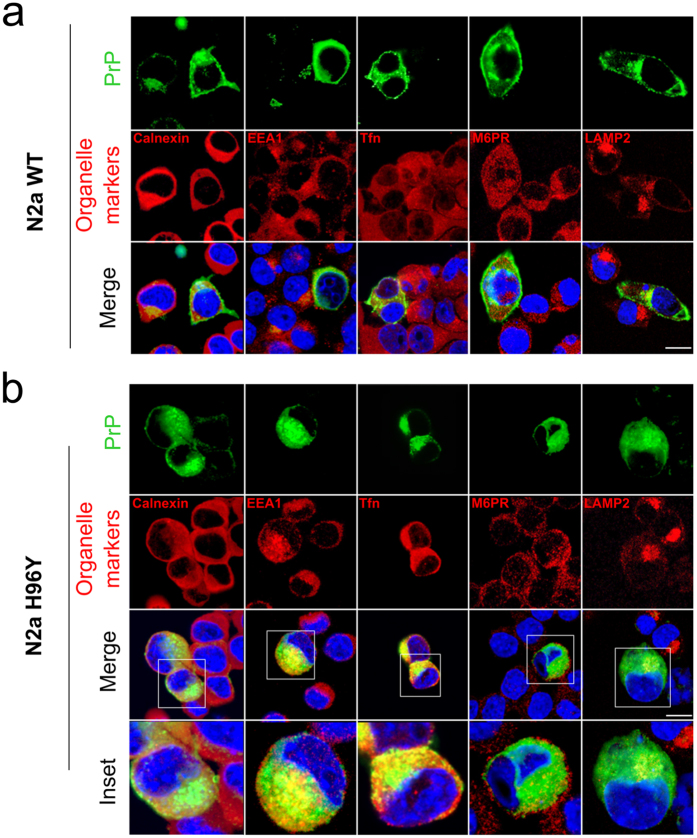
The observation that the H96Y mutant forms ThS-positive aggregates demands the identification of the primary intracellular compartments where H96Y mutant accumulation occurs. Consistent with previous studies, 3F4-WT MoPrP was found mostly on the cell surface and it was detectable along the ER, endosomal and lysosomal compartments. The WT and H96Y showed co-localization with the ER marker calnexin indicating a correct trafficking through the ER (Fig. 6a). Biochemical analysis on transfected WT and mutant MoPrP showed that the proteins display the same glycosylation patterns and molecular weight after Endo-H and PNGase-F treatments (Supplementary Fig. 2d). However, we found a significant population of H96Y mutant co-localizing with organelle markers as EEA1 (early endosomes), Tfn (recycling endosomes), M6PR (late endosome) and LAMP2 (lysosome marker) (Fig. 6b), suggesting a predominant accumulation of the mutant in the acidic compartments, as reported in previous studies on PrPSc intracellular accumulation.
Figure 6. The H96Y MoPrP mutant displays intracellular accumulation in the endosomial compartments.
PrP localization in N2a cells expressing the 3F4-WT MoPrP (a) or H96Y MoPrP (b). Nuclei are labeled with DAPI (blue), PrPs are detected by 3F4 antibody (green); organelle markers, such as Calnexin (ER marker), EEA1 (early endosomes marker), Tfn (recycling endosome marker), M6PR (late endosome marker) and LAMP2 (lysosome marker) are labeled in red. Insets in (b) shows a magnification of the merged panels (white boxed areas). Scale bars: 12 μm.
Removal of the H96-Cu(II) bond creates transient N-terminal β-sheet structures
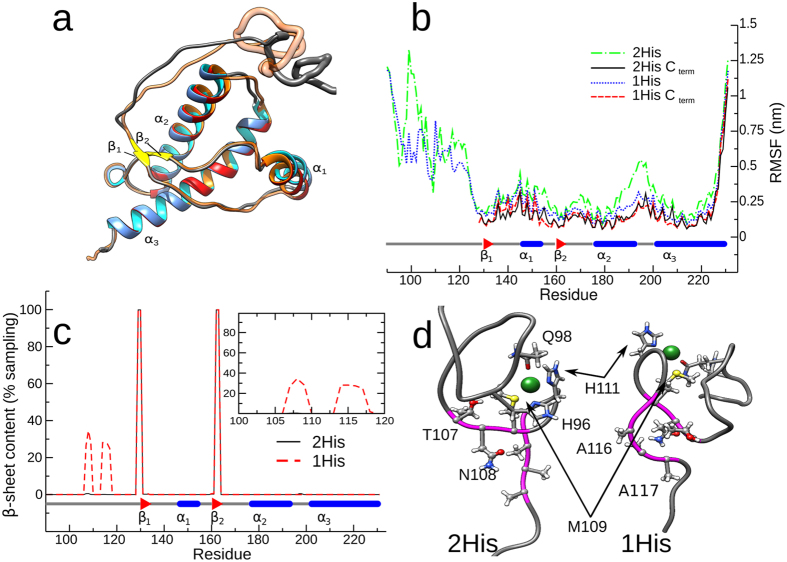
To interpret our results from a structural biology point of view, we performed MD simulation studies to predict the structural facets of WT HuPrP(90–231) coordinating Cu(II) either with His96 and His111 or with His111, termed 2His and 1His models, respectively.
The removal of H96 from the Cu(II) coordination sphere did not produce significant changes in the globular domain (residues 128–231), showing similar α2-α3 helix orientation as suggested by previous MD and NMR studies4,50 (e.g. α2-α3 helix angle is 51.2° and 49.1° in 2His and 1His models, respectively, while it is 50.9° in NMR structure) (Fig. 7a). While the C-terminal domain in the two simulation models featured a comparable flexibility (residue-wise Root Mean Square Fluctuation), the N-terminal segment of the 2His model displayed higher flexibility, particularly between residues 98–102 and 116–123 (Fig. 7b and Supplementary Fig. 3a). Additionally, the 2His trajectory yielded a slightly higher radius of gyration as compared to the 1His (1.77 ± 0.08 nm vs 1.74 ± 0.04 nm) with more pronounced oscillations (Supplementary Fig. 3b) confirming that the N-terminal segment in the 2His trajectory was relatively more disordered. The reduced flexibility of the residues 90–120 segment observed in the 1His trajectory is attributed to the presence of novel hydrogen (H)-bonds and salt bridges, which were uniquely present in the 1His simulation. In particular, we observed that in the 2His system the side-chain of T95 formed H-bond in 89% of sampling with the backbone oxygen of T107, the backbones of T95 and P105 formed a H-bond for 92% of sampling and the side-chains nitrogen atom of Q98 (a residue involved in copper binding) and N100 formed a H-bond in 51% of sampling. In the 1His trajectory, the backbones of residues N108 and A116 formed H-bond in 58% of sampling and the oxydryl group of T107 bound to the backbone of P105 in 62% of sampling. These alteration of the internal hydrogen bond network in the 1His system created favorable conditions for transient β-sheet motif formations in the segments formed by residues 106–109 and 114–117 (with a lifetime between 20% and 30% of sampling) which were not observed in the 2His trajectory (Fig. 7c and Supplementary Fig. 3a). A cluster analysis was performed on the N-terminal domain (residues 90–121) of the 2His and 1His trajectories, using 15000 even spaced configurations. More than 80% of the configurations in both trajectories were included in the first cluster (12017 for 2His and 13347 for 1His, respectively). Comparison of the first centroid structures obtained from the 1His and 2His simulations (Fig. 7d) shows that in the former case these groups of residues are indeed roughly antiparallel, with the side-chains of residues 106–109 oriented towards the backbone of residues 114–117, i.e. in a favorable position for the formation of a small β-sheet. On the other hand, in the 2His trajectory these two clusters form an angle of approximately 90° in most of the sampled structures.
Figure 7. The ion coordination sphere affects HuPrP(90–231) dynamics in MD simulations.
(a) Superimposition of average structures sampled in the 2His and 1His trajectories. Helical domains are represented in blue and red in 2His and 1His, respectively, the β1 and β2 sheets are in yellow, coils are represented in gray (2His) and orange (1His); the non-OR regions are depicted using a wider transparent ribbon. (b) Root Mean Square Fluctuation (RMSF) of residues for the 2His and 1His simulations. RMSF was calculated fitting the coordinates of the complete protein (2His: green dot-dashed line; 1His: blue dotted line) or restricted to the C-terminal domain (2His: black line; 1His: red dashed line). In both simulations the N-terminal domain is, as expected, much more flexible with the exception of residues 226–231; the 2His simulation apparently features greater fluctuations but the differences fade out when only the stable C terminal domain is considered. Notable exceptions are residues H140 and R151. (c) Residue-wise β-sheet content for the 2His and 1His simulation as a percentage of sampling (150.0 ns); the 1His simulation shows unstable β-sheet formation between residues 106–109 and 114–117 (also shown in the inset). (d) Comparison of the first centroid obtained by clustering the N-terminal domain Cα atoms in the 2His (left) and 1His (right) simulations; residues 106–109 and 114–117 are shown in ball and stick (backbone is shown in magenta as in Supplementary Fig 3a); copper binding residues H96, Q98 and H111 are shown as sticks and the copper ion as a dark green sphere (note that M109 belongs to both groups); a number of residues forming hydrogen bonds unique to one system are explicitly labeled to improve clarity.
Discussion
The central molecular event in prion diseases is the conversion of PrPC into pathological and infectious prions. Despite numerous investigations, the conversion mechanism(s) leading to PrPSc formation and transmission remain unclear. NMR-based studies on both Hu and MoPrP structured domains have proposed a role of the β2-α2 loop as dynamic “switch” element able to modulate prion conversion and susceptibility of a given specie to TSE51. However, the structural rearrangements occurring at the N-terminal region have not yet been clarified due to its intrinsic disorder. In a previous X-ray crystallography study we found that the palindromic domain can initiate β-sheet enrichment when HuPrP is crystallized in complex with a Nanobody24, suggesting that the segment 90–127 may act as alternative N-terminal switch for prion conversion. This region attracted interest because of the ability of the palindromic motif to form neurotoxic species52 and the proximity of the non-OR region, arguing a possible pathological link between copper binding to this site and prion conversion.
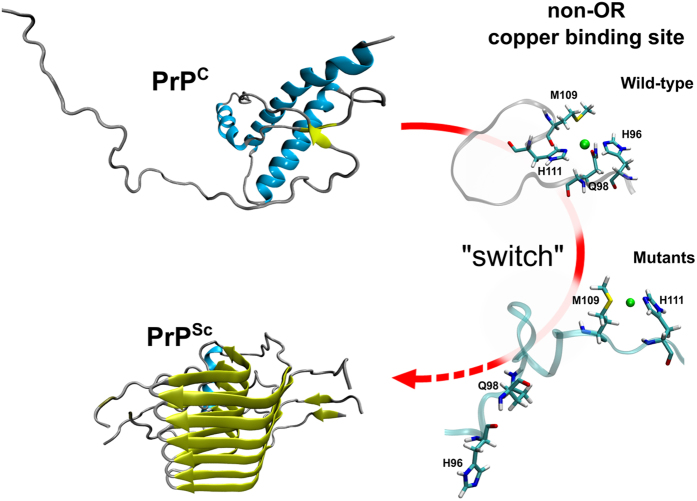
Here, we provide structural and biological evidence that the non-OR region may have pivotal biological implication for prion formation. We found that mutations –i.e. the GSS-causing Q212P and P102L mutants and the artificial H96Y mutation- and pH exchanges cause a dramatic modification on both Cu(II) and Cu(I) coordinations in the non-OR region. In the WT HuPrP Cu(II) and Cu(I) are anchored to His96 and His111 only at pH 5.5, while at pH 7 copper at both oxidative states is coordinated by His111. Conversely, in the mutants copper is bound only to H111 independently of the pH. The MD simulations propose a model whereby the molecular switch between the 2His and 1His systems (i.e. copper bound to H96 and H111, or only to H111, respectively) may have structural implications. The 1His coordination seems to favor β-sheet enrichment in the region encompassing residues 106–117. At this stage it is unclear how this early β-sheet conversion may drive a complete structural conversion to PrPSc, but the new structural rearrangements occurring at the palidromic motif might serve as a nucleus for the association of intermolecular β-strands.
To interpret these structural clues in the context of more relevant physiological implications, we used ScN2a and N2a cells transiently expressing 3F4-MoPrP with the H96Y mutation. This construct mimics the pathological P102L and Q212P mutants investigated by EXFAS and it may represent a useful model to elucidate the role of the non-OR region for prion conversion. His to Tyr96 substitution removes one crucial copper ligand, thus it may allow to link the effect caused by altered copper coordination with prion conversion. The expression of H96Y mutant highly promotes prion conversion in ScN2a cells and spontaneously generated PrPres together with intracellular MoPrP accumulation in N2a cells. We hypothesize that the non-OR copper-binding site at H96 is much more important for both PrPC function, as shown by the higher affinity for copper binding compared to the OR region53, and prion propagation and infectivity54. Residue H96 is located in a PrPSc region that is partially resistant to PK, thus copper bound to H96 may have a role during PrPSc formation even in the absence of the OR region55. On the basis of our findings, we propose that the H96Y mutant may act as other pathogenic mutations located in both N- and C-terminal domains (e.g. P102L and Q212P), causing spontaneous prion conversion. Our analysis shows that substitution of H96Y mutation induces PrPC accumulation in the acidic endosomal compartments, as observed for other disease-linked mutations56. The pH change from neutral to acidic values has been proposed to trigger the PrPC conformational conversion and a change in the balance of distinct internalization mechanisms may promote PrPSc replication by diverting the protein to distinct intracellular compartments and inhibiting the cellular protein quality control systems57. We report here enhanced PK-resistance of the H96Y mutant when expressed in ScN2a cells and its ability to generate de novo PrPres in N2a cells. These findings propose a pivotal role for non-OR region as critical molecular switch for prion conversion. We therefore argue that copper bound to the non-OR region may stabilize this segment when coordinated by His96 and His111, preventing misfolding events through transient short and long range interaction contacts between the 90–127 residues and the C-terminal structured domain. Copper acts as a key modulator of this process since its absence from PrPC side promote prion conversion, as observed in WT ScN2a cells treated with a copper chelator.
Our study highlights the importance of the non-OR region for prion conversion and suggests a model in which PrPC coordinating copper with one His may be more prone to the conversion at acidic condition (Fig. 8). Thus, the non-OR region may act as the long-sought-after prion switch. The data presented here provide the bases for experiments aimed at elucidating whether the H96Y mutation causes TSE when expressed in animal models.
Figure 8. Model for the non-OR region molecular switch at acidic pH.
PrPC coordinating copper with one His residue in the non-OR region is more prone to the conversion at acidic pH condition. We propose a model where His96 and His111 represent the N-terminal switch for prion conversion in the PrPC. As PrPSc model we used the HET-s(218–289) 2KJ3 PDB structure.
Additional Information
How to cite this article: Giachin, G. et al. The non-octarepeat copper binding site of the prion protein is a key regulator of prion conversion. Sci. Rep. 5, 15253; doi: 10.1038/srep15253 (2015).
Supplementary Material
Acknowledgments
This work was supported by European Community’s Seventh Framework Programme (FP7 grant agreement no. 222887 – the PRIORITY project, to G.L.) and by the University of Rome “La Sapienza” (Progetto Ateneo 2014, n.C26A14L7CX, to P.D.). We thank Dr Andrea Zitolo for initial work on EXAFS data collection. We are thankful the European Synchrotron Radiation Facility and Isabelle Kieffer at BM30B FAME for the invaluable support.
Footnotes
Author Contributions G.G. and P.T.M. contributed equally to this work. P.D. and G.L. conceived the project and jointly supervised this work. G.G., G.M., P.D. and G.L. wrote the manuscript. G.G., A.A. and P.D. carried out EXAFS data collection at ESRF. G.G., V.M. and P.D. analyzed EXAFS data. P.T.M. carried out the in vitro and cell-based assays. G.G. and P.T.M. analyzed the in vitro and cell-based data. G.M. carried out and analyzed the molecular dynamic experiments. G.S. and T.H.T. contributed to the recombinant proteins production. T.H.T. contributed to cuprizone treatment experiments. F.B. and S.D. contributed to the discussion of the results. All authors read and approved the final manuscript.
References
- Colby D. W. & Prusiner S. B. Prions. Cold Spring Harb Perspect Biol 3, a006833 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legname G., Giachin G. & Benetti F. Structural Studies of Prion Proteins and Prions. In: Non-fibrillar Amyloidogenic Protein Assemblies - Common Cytotoxins Underlying Degenerative Diseases (ed^(eds Rahimi F. & Bitan G.) Springer: Netherlands, (2012). [Google Scholar]
- Surewicz W. K. & Apostol M. I. Prion protein and its conformational conversion: a structural perspective. Top Curr Chem 305, 135–167 (2011). [DOI] [PubMed] [Google Scholar]
- Zahn R. et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci USA 97, 145–150 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter E. D., Chattopadhyay M. & Millhauser G. L. The affinity of copper binding to the prion protein octarepeat domain: evidence for negative cooperativity. Biochemistry 45, 13083–13092 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter E. D. et al. Copper binding extrinsic to the octarepeat region in the prion protein. Curr Protein Pept Sci 10, 529–535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling M. F. et al. The hydrophobic core sequence modulates the neurotoxic and secondary structure properties of the prion peptide 106-126. J Neurochem 73, 1557–1565 (1999). [DOI] [PubMed] [Google Scholar]
- Gong B. et al. Probing structural differences between PrP(C) and PrP(Sc) by surface nitration and acetylation: evidence of conformational change in the C-terminus. Biochemistry 50, 4963–4972 (2011). [DOI] [PubMed] [Google Scholar]
- Requena J. R. & Wille H. The structure of the infectious prion protein. Prion 8, 60–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuvolone M., Aguzzi A. & Heikenwalder M. Cells and prions: a license to replicate. FEBS Lett 583, 2674–2684 (2009). [DOI] [PubMed] [Google Scholar]
- Herms J. et al. Evidence of presynaptic location and function of the prion protein. J Neurosci 19, 8866–8875 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A. D., Emsley J. G., Ozdinler P. H., Lindquist S. & Macklis J. D. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc Natl Acad Sci USA 103, 3416–3421 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiati M. D. et al. PrPC controls via protein kinase A the direction of synaptic plasticity in the immature hippocampus. J Neurosci 33, 2973–2983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuccione A., Sytnyk V., Leshchyns’ka I. & Schachner M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J Cell Biol 169, 341–354 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini L., Meneghetti E., Pastore B., Benetti F. & Legname G. Prion Protein and Copper Cooperatively Protect Neurons by Modulating NMDA Receptor Through S-nitrosylation. Antioxidants & Redox Signaling 22, 772–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravani H. et al. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J Cell Biol 181, 551–565 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stys P. K., You H. & Zamponi G. W. Copper-dependent regulation of NMDA receptors by cellular prion protein: implications for neurodegenerative disorders. J Physiol 590, 1357–1368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushie M. J. et al. Prion protein expression level alters regional copper, iron and zinc content in the mouse brain. Metallomics 3, 206–214 (2011). [DOI] [PubMed] [Google Scholar]
- Brown D. R. Prions and manganese: A maddening beast. Metallomics 3, 229–238 (2011). [DOI] [PubMed] [Google Scholar]
- Walter E. D., Stevens D. J., Visconte M. P. & Millhauser G. L. The prion protein is a combined zinc and copper binding protein: Zn2+ alters the distribution of Cu2+ coordination modes. J Am Chem Soc 129, 15440–15441 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos J., Kovacs I., Hajos F., Kalman M. & Simonyi M. Nerve endings from rat brain tissue release copper upon depolarization. A possible role in regulating neuronal excitability. Neurosci Lett 103, 139–144 (1989). [DOI] [PubMed] [Google Scholar]
- Vassallo N. & Herms J. Cellular prion protein function in copper homeostasis and redox signalling at the synapse. J Neurochem 86, 538–544 (2003). [DOI] [PubMed] [Google Scholar]
- Fischer M. et al. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J 15, 1255–1264 (1996). [PMC free article] [PubMed] [Google Scholar]
- Abskharon R. N. et al. Probing the N-terminal beta-sheet conversion in the crystal structure of the human prion protein bound to a nanobody. J Am Chem Soc 136, 937–944 (2014). [DOI] [PubMed] [Google Scholar]
- Younan N. D. et al. Copper(II)-induced secondary structure changes and reduced folding stability of the prion protein. J Mol Biol 410, 369–382 (2011). [DOI] [PubMed] [Google Scholar]
- Migliorini C., Sinicropi A., Kozlowski H., Luczkowski M. & Valensin D. Copper-induced structural propensities of the amyloidogenic region of human prion protein. J Biol Inorg Chem 19, 635–645 (2014). [DOI] [PubMed] [Google Scholar]
- Baumann F. et al. Lethal recessive myelin toxicity of prion protein lacking its central domain. EMBO J 26, 538–547 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. et al. Neonatal lethality in transgenic mice expressing prion protein with a deletion of residues 105-125. EMBO J 26, 548–558 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald A., Pushie M. J., Millhauser G. L. & George G. N. New insights into metal interactions with the prion protein: EXAFS analysis and structure calculations of copper binding to a single octarepeat from the prion protein. J Phys Chem B 117, 13822–13841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo P. et al. Effects of the pathological Q212P mutation on human prion protein non-octarepeat copper-binding site. Biochemistry 51, 6068–6079 (2012). [DOI] [PubMed] [Google Scholar]
- Hasnain S. S. et al. XAFS study of the high-affinity copper-binding site of human PrP(91-231) and its low-resolution structure in solution. J Mol Biol 311, 467–473 (2001). [DOI] [PubMed] [Google Scholar]
- Ilc G. et al. NMR structure of the human prion protein with the pathological Q212P mutation reveals unique structural features. PLoS One 5, e11715 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J. R., Grinstein S. & Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol 11, 50–61 (2010). [DOI] [PubMed] [Google Scholar]
- Parchi P. et al. Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann-Straussler-Scheinker disease. Proc Natl Acad Sci USA 95, 8322–8327 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proux O. et al. FAME: A new beamline for X-ray absorption investigations of very-diluted systems of environmental, material and biological interests. Phys Scripta T115, 970–973 (2005). [Google Scholar]
- Filipponi A., Di Cicco A. & Natoli C. R. X-ray-absorption spectroscopy and n-body distribution functions in condensed matter. I. Theory. Phys Rev B Condens Matter 52, 15122–15134 (1995). [DOI] [PubMed] [Google Scholar]
- Filipponi A. & Di Cicco A. X-ray-absorption spectroscopy and n-body distribution functions in condensed matter. II. Data analysis and applications. Phys Rev B Condens Matter 52, 15135–15149 (1995). [DOI] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. C. et al. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 33, W368–371 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose F., Hodak M. & Bernholc J. Mechanism of copper(II)-induced misfolding of Parkinson’s disease protein. Sci Rep 1, 11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliev A. E. et al. Motional timescale predictions by molecular dynamics simulations: case study using proline and hydroxyproline sidechain dynamics. Proteins 82, 195–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D. et al. GROMACS: fast, flexible, and free. J Comput Chem 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics 79, 926–935 (1983). [Google Scholar]
- Cheatham T. E., Miller J. L., Fox T., Darden T. A. & Kollman P. A. Molecular-Dynamics Simulations on Solvated Biomolecular Systems - the Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, Rna, and Proteins. Journal of the American Chemical Society 117, 4193–4194 (1995). [Google Scholar]
- Hess B., Bekker H., Berendsen H. J. C. & Fraaije J. G. E. M. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry 18, 1463–1472 (1997). [Google Scholar]
- Bussi G., Donadio D. & Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- Torda A. E. & Vangunsteren W. F. Algorithms for Clustering Molecular-Dynamics Configurations. Journal of Computational Chemistry 15, 1331–1340 (1994). [Google Scholar]
- Kabsch W. & Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- Benetti F. et al. Cuprizone neurotoxicity, copper deficiency and neurodegeneration. Neurotoxicology 31, 509–517 (2010). [DOI] [PubMed] [Google Scholar]
- Rossetti G., Giachin G., Legname G. & Carloni P. Structural facets of disease-linked human prion protein mutants: a molecular dynamic study. Proteins 78, 3270–3280 (2010). [DOI] [PubMed] [Google Scholar]
- Sigurdson C. J. et al. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest 120, 2590–2599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling M. F. et al. Copper and zinc binding modulates the aggregation and neurotoxic properties of the prion peptide PrP106-126. Biochemistry 40, 8073–8084 (2001). [DOI] [PubMed] [Google Scholar]
- Nadal R. C., Davies P., Brown D. R. & Viles J. H. Evaluation of copper2+ affinities for the prion protein. Biochemistry 48, 8929–8931 (2009). [DOI] [PubMed] [Google Scholar]
- Quaglio E., Chiesa R. & Harris D. A. Copper converts the cellular prion protein into a protease-resistant species that is distinct from the scrapie isoform. J Biol Chem 276, 11432–11438 (2001). [DOI] [PubMed] [Google Scholar]
- Cox D. L., Pan J. & Singh R. R. A mechanism for copper inhibition of infectious prion conversion. Biophys J 91, L11–13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok A. & Hegde R. S. Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathog 5, e1000479 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskakov I. V., Legname G., Baldwin M. A., Prusiner S. B. & Cohen F. E. Pathway complexity of prion protein assembly into amyloid. J Biol Chem 277, 21140–21148 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.