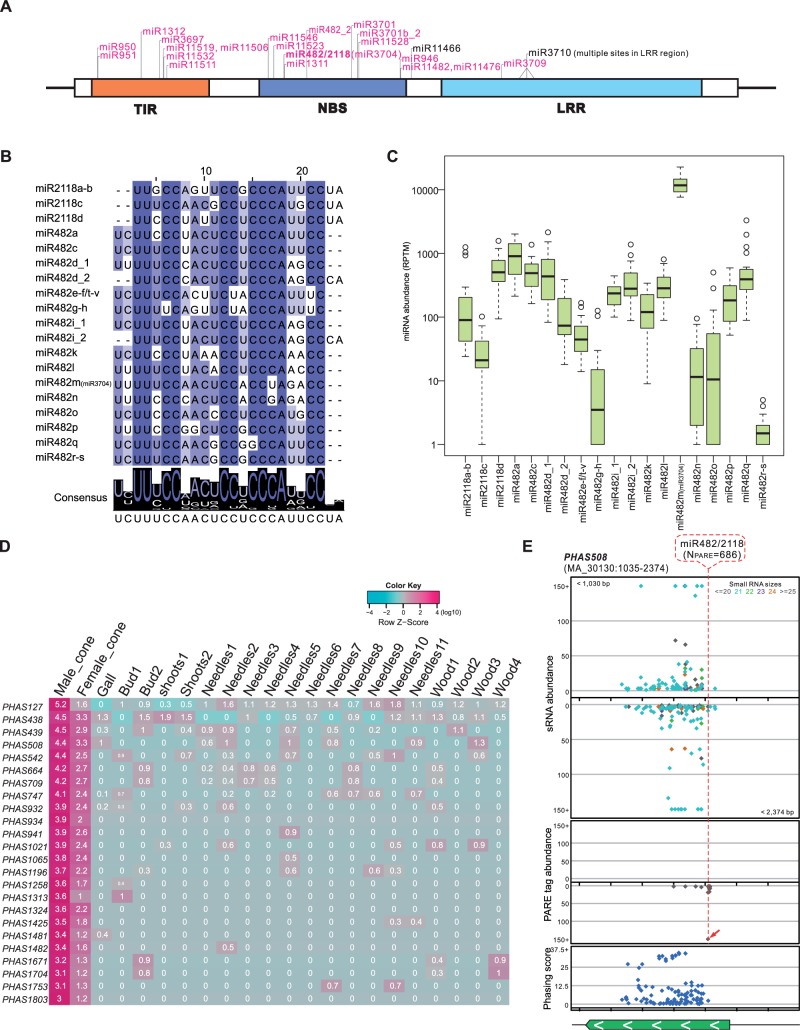
Fig. 2.
An expanded miRNA-NB-LRR-phasiRNA network in Norway spruce. (A) Target site distribution of miRNAs identified in spruce targeting NB-LRRs. A prototypical NB-LRR gene encodes three conserved domains: TIR (Toll/interleukin receptor-like domain), NBS, and an LRR (leucine-rich repeat) region. The 22-nt miRNAs are marked in pink. Multiple target sites for miR3710 (21 nt) are found in the region coding for the LRR domain. (B) Alignment of mature sequences for all members of the miR482/miR2118 superfamily in spruce. The degree of conservation for each nucleotide along the miRNAs is represented by the color, with a dark color denoting a high level of conservation and a light color denoting a low level. The consensus sequence of the alignment is displayed below with sequence logos. (C) Abundance of miR482/miR2118 mature sequences in 22 tissues of spruce. (D) PhasiRNAs of 24 noncoding PHAS loci specifically accumulated in reproductive tissues. Values are displayed in a log10 scale, with the pink color denoting high abundance and green indicating low abundance. Column titles indicate their source tissues, as detailed in supplementary table S1, Supplementary Material online. (E) An example of a noncoding PHAS locus (PHAS508) targeted by miR482/miR2118. Distributions of sRNAs, PARE data, and phasing score of the PHAS508 loci are viewed in different tracks. Dots in different colors in the sRNA track denote sRNAs of different lengths. A transcript annotated in the spruce genome (http://congenie.org/gbrowse) is shown at the bottom with the transcription direction indicated with white arrows. Introns in a gene are indicated by light-orange boxes.