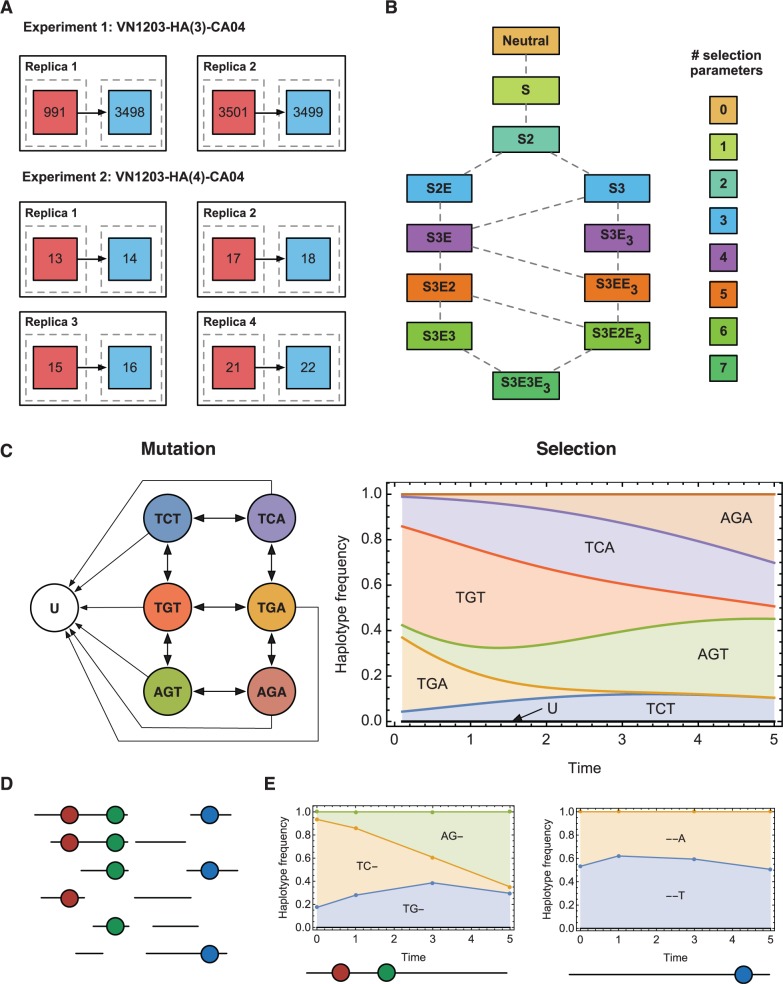
Fig. 1.
Overview of the method. (A) Original experimental setup. A large dose of virus was given to a ferret (red), contact (via airborne transmission) with a second ferret (blue) being initiated 1 day later. Two different viral strains were used; sequence data from two and four replicate experiments being described in each case. (B) Network of potential fitness landscape models for an example three-locus haplotype system. Here, S indicates an allele under constant selection, E indicates a two-locus epistatic interaction between alleles, and E3 indicates a three-locus epistatic interaction between alleles. Each model represents multiple potential fitness landscapes according to the choice of selected alleles and selection coefficients; model S encompasses all landscapes with an allele at any one locus under selection of any strength. (C) Dynamics in a single model. Mutation occurs between observed haplotypes differing by a single allele, and to a class U, representing unobserved haplotypes. Under selection, the frequencies of the different haplotypes change over time. (D) Paired-end sequence reads span either one or multiple alleles of interest in the system. Reads are partitioned according to the alleles they span. (E) Sets of reads describe partial haplotype frequencies, to which evolutionary models are fitted.