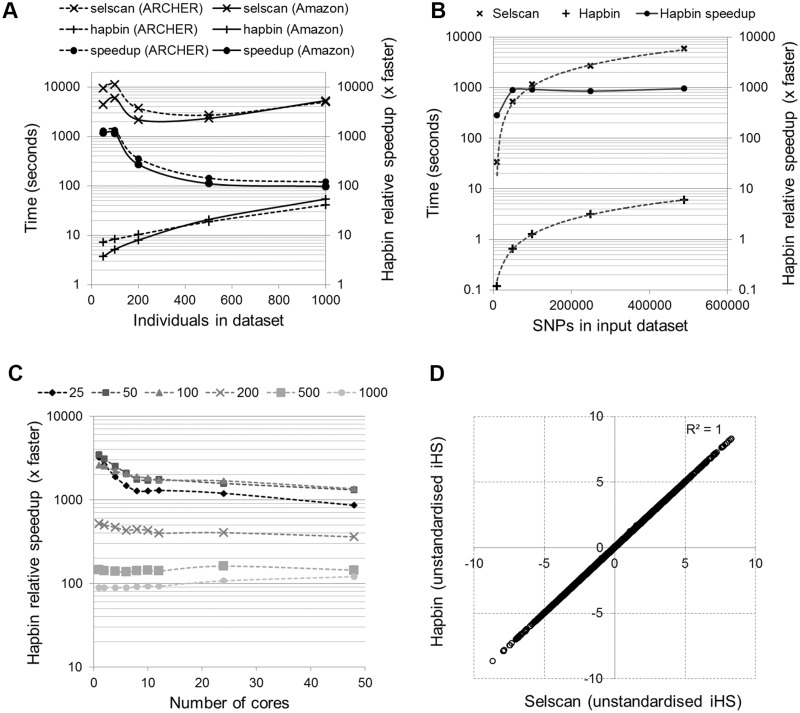
Fig. 1.
Hapbin versus selscan comparisons. (A) Time taken by hapbin and selscan to calculate iHS across chromosome 22 across 48 cores (1 node) onz ARCHER and on an Amazon c3.8xlarge instance. Subsets of individuals being randomly sampled from the 1000 genomes dataset. (B) Time taken by hapbin and selscan to calculate iHS in the 1000 genomes GBR (Great Britain) population of 89 individuals on the Amazon c3.8xlarge instance. Runs of contiguous SNPs by location were subsampled from all of those on chromosome 22. (C) Hapbin’s relative speedup versus selscan when run across chromosome 22 with varying numbers of cores and individuals on ARCHER. (D) Comparison of unstandardized iHS values output by both selscan and hapbin when run across 500 randomly selected individuals and all SNPs on chromosome 22.