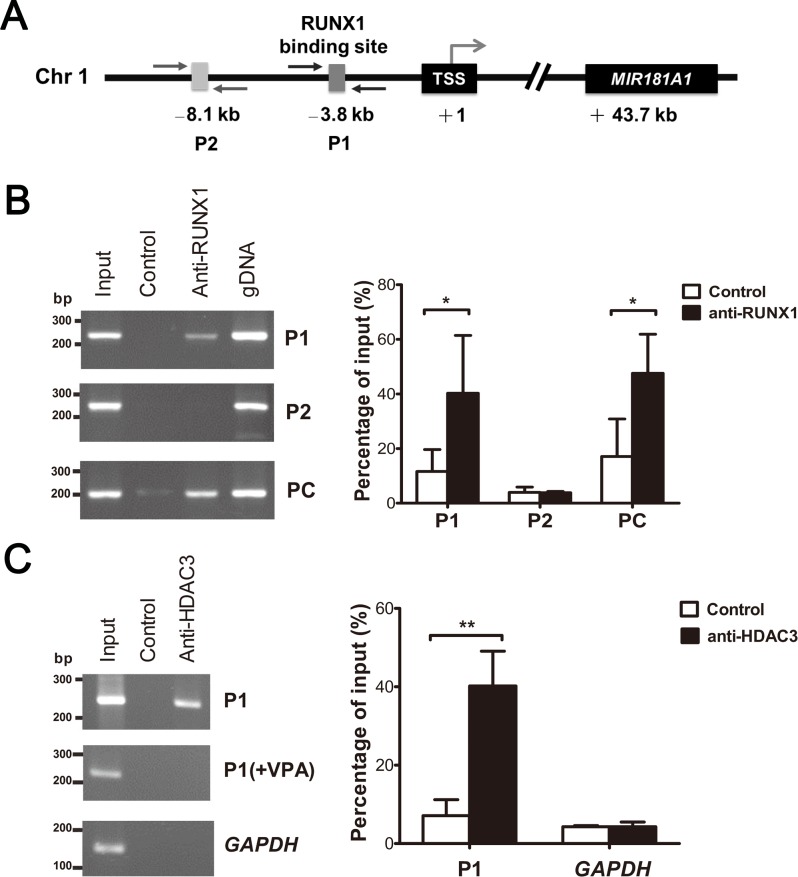
Fig 2. ETV6/RUNX1 inhibits MIR181A1 transcription via binding to the endogenous RUNX1 site.
(A) Schematic representation of the genomic structure of human MIR181A1. The location of the MIR181A1 gene and the RUNX1-binding site are numbered relative to the TSS (+1). Arrows indicate the locations of the primers used in the ChIP assay. (B) ChIP was carried out using anti-RUNX1 or in the absence of specific antibody (Control) (left). DNA sequences surrounding the putative RUNX1-binding site were amplified by PCR using P1 primers. To evaluate the specificity of RUNX1 binding, a positive control and a negative control were performed using PC and P2, respectively, for the ChIP assay. Amplification of the upstream region near the RUNX1-binding site on MIR223, which is a known direct target of RUNX1, was performed using PC primers. P2 primers were designed to amplify a distal region lacking the RUNX1-binding site. Input shows the amplification from sonicated chromatin, and genomic DNA (gDNA) was used as a positive PCR control. The PCR products were quantified by densitometry (right). (C) The ChIP assay was performed using anti-HDAC3 (left). Treatment with valproic acid and amplification of the promoter region of GAPDH were used as controls. The PCR products were quantified by densitometry (right). Bars show the mean ± SD from three independent experiments. *P ≤ 0.05, **P ≤ 0.01 (ANOVA).