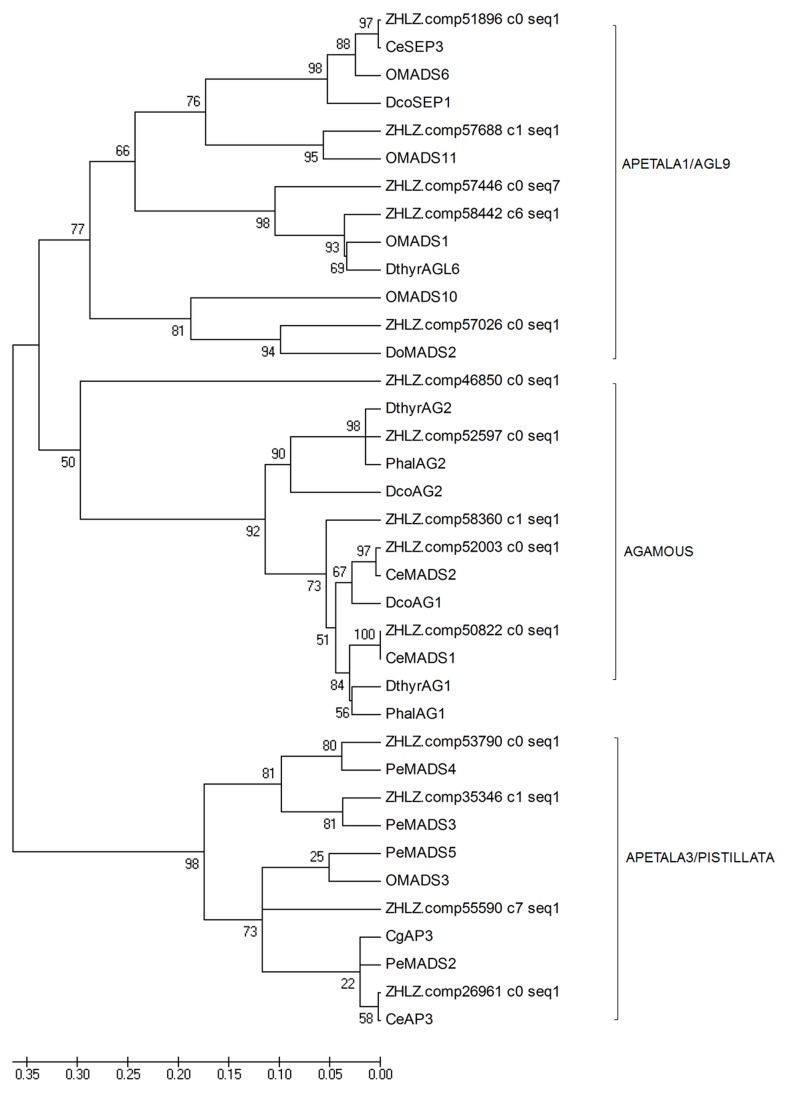
Fig 12. Phylogenetic analysis of the MADS-box genes differentially expressed among different floral organs.
Amino acid sequences were aligned by the ClustalW 2.0, and phylogenetic relationships were reconstructed using a maximum-likelihood (ML) method in PHYML software with JTT amino acid substitution model. Previously published plant MADS-box protein sequences were retrieved from GenBank database (PeMADS2: AAR26628, CeAP3-like1: AFH66788, CeAP3-like2: AFH66787, CgAP3:ADI58460, CeMADS2: ADP00516, CeMADS1: ADP00515, CePI-like: AFH66786, OMADS11: ADJ6724, OMADS6: ADJ67238, OMADS1: ADJ67237, OMADS10: ADJ67240, OMADS2: AIJ29175, OMADS3: AAO45824, OMADS4: AIJ29176, DthyrAG1: AAY86364, DthyrAG2: AAY86365, PhalAG1: BAE80120, PhalAG2: BAE80121, PeMADS2: AY378149, PeMADS3: AY378150, PeMADS4: AY378151, PeMADS5: AY378148).