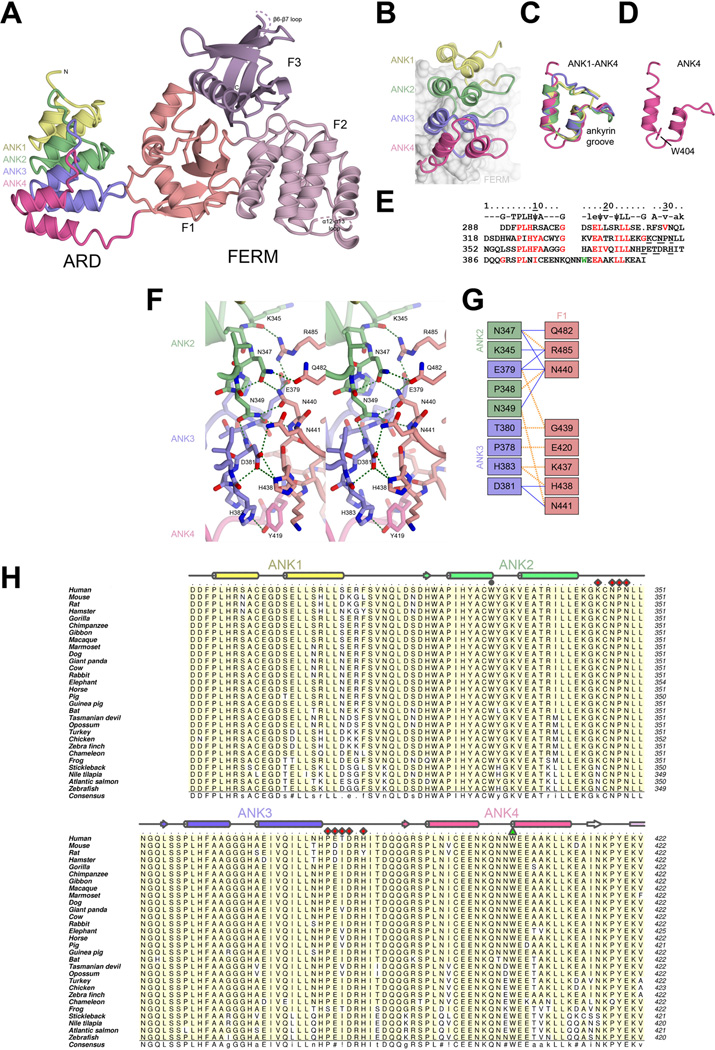
Figure 2. Structure of KRIT1 ARD-FERM.
A) Overall structure of KRIT1 ARD-FERM shown in cartoon format. ARD is colored showing each ankyrin repeat (ANK1–ANK4). FERM domain colored as per Figure 1. B–E) Analysis of the KRIT1 ARD. B) Close-up of the ARD showing each ankyrin repeat. The FERM domain is shown as a surface in the background. C) Superposition of each of the ankyrin repeats. D) The fourth ankyrin repeat alone, showing residue W404 which stabilizes an insert (Q401, N402, N403). E) Alignment of KRIT1 ankyrin repeats. Top row shows consensus ankyrin repeat sequence with well conserved ankyrin repeat residues indicated by capitals, moderately conserved ankyrin repeat residues in lower case, and hydrophobic residues in the consensus indicated by ψ (Mosavi et al., 2004; Sedgwick and Smerdon, 1999). KRIT1 residues that conform to the consensus are colored red. W404 is colored green. Residues that interact with the FERM domain are underlined. F) Stereoview of the intramolecular interaction between KRIT1 ARD and FERM domains. ARD ankyrin repeats colored as in panel B, FERM F1 colored as in panel A. Hydrogen bonds are indicated in green. Interacting residues are shown in stick format with oxygen atoms colored red and nitrogen atoms colored blue. G) Schematic map of the ARD-FERM interface. Hydrophobic interactions are indicated by orange dashed lines and hydrogen bonds by solid blue lines. H) Sequence alignment of KRIT1 ARD. Secondary structure elements colored per ankyrin repeat with α-helices shown as cylinders and β-strands as arrows. ARD residues that interact with KRIT1 FERM domain are indicated with red diamonds, and W404 is shown with a green triangle. W330 is indicated with a grey circle. Uniprot or GenBank accession number are followed by the Latin name and label in parentheses: O00522 Homo sapiens (Human), Q6S5J6 Mus musculus (Mouse), B5DF47 Rattus norvegicus (Rat), XP_003496996 Cricetulus griseus (Hamster), G3QHP7 Gorilla gorilla (Gorilla), XP_001166592 Pan troglodytes (Vhimpanzee), G1RZ72 Nomascus leucogenys (Gibbon), F6TVZ5 Macaca mulatta (Macaque), F7I3T9 Callithrix jacchus (Marmoset), E2RAA5 Canis familiaris (Dog), XP_002925462 Ailuropoda melanoleuca (Giant panda), Q6TNJ1 Bos taurus (Cow), G1SM20 Oryctolagus cuniculus (Rabbit), G3TAH0 Loxodonta africana (Elephant), XP_001491579 Equus caballus (Horse), F1SFD0 Sus scrofa (Pig), XP_003475143 Cavia porcellus (Guinea pig), G1PWA1 Myotis lucifugus (Bat), G3X3N7 Sarcophilus harrisii (Tasmanian devil), F7CJF5 Monodelphis domestica (Opossum), G1N921 Meleagris gallopavo (Turkey), NP_001026144 Gallus gallus (Chicken), XP_002196126 Taeniopygia guttata (Zebra finch), G1KRC6 Anolis carolinensis (Chameleon), F7CJC5 Xenopus tropicalis (Frog), G3NJI7 Gasterosteus aculeatus (Stickleback), XP_003452511 Oreochromis niloticus (Nile tilapia), C0H9F0 Salmo salar (Atlantic salmon), B8JIZ5 Danio rerio (Zebrafish). Alignment was conducted using ClustalW (Larkin et al., 2007). Figure was generated using Aline (Bond and Schuttelkopf, 2009). Interacting residues were defined using the PISA server (Krissinel and Henrick, 2007).