Abstract
Proper DNA replication is critical to maintain genome stability. When the DNA replication machinery encounters obstacles to replication, replication forks stall and the replication stress response is activated. This response includes activation of cell cycle checkpoints, stabilization of the replication fork, and DNA damage repair and tolerance mechanisms. Defects in the replication stress response can result in alterations to the DNA sequence causing changes in protein function and expression, ultimately leading to disease states such as cancer. To identify additional genes that control the replication stress response, we performed a three-parameter, high content, whole genome siRNA screen measuring DNA replication before and after a challenge with replication stress as well as a marker of checkpoint kinase signalling. We identified over 200 replication stress response genes and subsequently analysed how they influence cellular viability in response to replication stress. These data will serve as a useful resource for understanding the replication stress response.
Keywords: replication stress, ATR, checkpoint, RNAi screen, DNA damage, hydroxyurea, gemcitabine, camptothecin, PARP inhibitor
1. INTRODUCTION
As the replication machinery traverses the DNA, obstacles created by both exogenous and endogenous sources can impede the replication fork causing replication stress (1–3). Sources of replication stress include DNA damage, DNA structures that pose a physical barrier to fork progression, nicks and gaps formed as intermediates of DNA repair, the collision of replication forks with transcription machinery, and limiting amounts of nucleotides or other replication factors (3). When the replication machinery encounters these obstructions it slows or stalls, and frequently the DNA polymerase becomes uncoupled from the replicative helicase (4). Uncoupling results in tracts of single-stranded DNA (ssDNA) that are subsequently coated by the ssDNA binding protein replication protein A (RPA). ATRIP binds RPA, recruiting the ATR kinase to the site of replication stress (5–7). ATR inhibits cell cycle progression, promotes damage repair, and helps to stabilize and restart the fork by phosphorylating and activating proteins that function in these processes (8).
Unresolved DNA damage and replication stress can result in DNA alterations ultimately leading to diseases including cancer. Indeed, defects in replication stress response proteins cause Schimke immuno-osseus dysplasia, Seckel, Bloom, Rothmund-Thomson, and Werner syndromes (3). Additionally, many cancer cells contain high replication stress levels due to activated oncogenes and/or defects in genome maintenance activities (9–11). Combined with the frequent loss of the p53-dependent G1 checkpoint and apoptotic pathways, the high levels of replication-associated damage in cancer cells create an increased dependence on the replication stress response for continued proliferation and viability. Thus, the replication stress response has become a focus of interest for targeted cancer therapies.
To better understand the mechanisms promoting DNA synthesis and the maintenance of genome integrity in cells we completed a screen to identify replication stress response genes. The whole-genome siRNA screen utilized immunofluorescent measurements of DNA synthesis before and after a replication stress challenge as well as a measurement of replication stress response signalling. Over 200 replication stress response genes, including known and novel genes, were identified which will serve as a useful resource for future investigation.
2. MATERIAL AND METHODS
Cell Culture
U2OS cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 7.5% fetal bovine serum (FBS).
Primary and Validation Screens
The screens were performed in triplicate in 384-well plates containing sample, All Stars Negative Control (non-targeting) (Qiagen), ATR positive control (Dharmacon), and cell death transfection efficiency control (Qiagen) siRNAs. The primary screen utilized a whole genome siRNA library (Dharmacon ON-TARGETplus SMARTpool siRNA library – Human Drug targets, Human Druggable Subsets, and Human Genome) consisting of a pool of 4 siRNAs per gene. A total of 18,055 genes were screened. The validation screen utilized a custom siRNA library consisting of 4 individual siRNAs per gene (Dharmacon siGENOME siRNAs). 10nM siRNA was transfected into U2OS cells using Dharmafect 1 transfection reagent (Dharmacon). Seventy-two hours post-transfection, cells were incubated in media containing 10µM BrdU for 30 minutes, washed, and treated with 2mM HU for 24 hours. Cells were subsequently washed to remove the HU, and labelled with 10µM EdU for 4 hours prior to fixing with 3.7% paraformaldehyde in PBS. Samples were permeabilized with 0.5% Triton-X100 in PBS for 20 minutes, washed with PBS, and incubated in EdU Click-iT reaction buffer (1:200 AlexaFluor Azide 488 (Life Technologies), 2mg/ml sodium ascorbate, and 1.5mM copper sulfate in PBS) for 30 minutes. After washing again with PBS, the cells were treated for 30 minutes with 10% Image-iT FX Signal Enhancer (Life Technologies) in PBS, washed, and blocked with 10% FBS in PBS for 30 minutes. Samples were incubated overnight at 4°C in primary antibody (1:60 MOBU-1 BrdU antibody (Life Technologies), 1:1000 Benzonase (Novagen), and 1:9000 γH2AX antibody (Bethyl, custom) in 1% BSA in PBS). The following day, samples were washed with PBS, incubated for 20 minutes in secondary antibody (1:800 Cy5 (Life Technologies) and 1:500 AlexaFluor 568 (Life Technologies) in 1% BSA in PBS), washed with PBS, and labelled with 200ng/ml DAPI in PBS for 3 minutes.
Immunofluorescent images were obtained using the Perkin Elmer Opera QEHS system. Images at six different locations were taken per well and analysed with Perkin Elmer’s Columbus software. Individual nuclei were identified utilizing DAPI, nuclei on the border of images removed, and intensity levels of BrdU, γH2AX, and EdU determined for each nucleus. The ratio of the γH2AX/EdU intensity was calculated per nucleus, and the mean calculated to obtain a replication restart score (RRS) for each sample. Robust Z-scores ((x-median)/(MAD*1.4826)) for the RRS, γH2AX, and EdU were calculated for each sample on a plate-by-plate basis. The robust Z-score allows for normalization of data to provide information on the strength of each siRNA in relation to all samples within a plate utilizing the median and median absolute deviation, which are less sensitive to outliers than mean values. The sum of the Z-scores for three replicates was determined to obtain one value by which to analyse each gene. 530 genes had an RRS Z-score sum greater than or equal to 15. Genes that were essential for viability were discarded (<60 cells in all three replicates). The 200 genes with the highest RRS robust Z-score sum and those detected in other published screens analysing genome integrity and the phosphoproteome of ATM and ATR (12–23) were selected for further analysis in the validation and secondary screens.
The validation screen was performed using the same procedure as the primary screen. RRS were determined for each nucleus and the RRS from 3 replicate samples compared to non-targeting controls by the Wilcoxon test. The most statistically significant genes were selected using the B statistic (the log odds that the sample siRNA is differentially expressed) >1 and a false-discovery rate (FDR)-adjusted p-value ≤ 0.01 (24).
Drug Sensitivity Screens
384-well plates containing sample, non-targeting, ATR, and cell death siRNAs were utilized. 10nM siRNA was transfected into U2OS cells using Dharmafect 1 transfection reagent. Seventy-two hours post-transfection, cells were split 1:4 into four 384-well plates and either mock-treated or treated with 0.2mM HU, 0.1µM ATR inhibitor (VX970), 0.05µM CHK inhibitor (AZD7762), 1nM gemcitabine, 5nM camptothecin, or 10nM PARP inhibitor (BMN673) for 72 hours. Cells were then washed, incubated in media containing alamar blue (Life Technologies) for 4 hours and 595nM absorbance readings obtained. The viability screens were performed in triplicate.
Data were first corrected by subtracting background absorbance values. The viability ratio for each siRNA was calculated by dividing the alamar blue absorbance on the drug treatment plate by the absorbance on the mock treated plate. These viability ratios were then normalized for plate-to-plate variation by dividing by the average ratio of untreated non-targeting controls on drug treatment/mock treatment for each plate. The normalized viability ratios were log2 transformed and each siRNA compared to the non-targeting control by two-tailed t-test using a FDR-adjusted significance level of 1 percent by the Benjamini and Hochberg method. siRNAs with normalized viability ratios of at least 15 percent below non-targeting control samples for each drug treatment with an FDR-adjusted p-value of 0.01 or lower were identified as causing hypersensitivity.
GESS Analysis
Potential siRNA off-target effects were determined utilizing the Genome-wide Enrichment for Seed Sequence match (GESS) analysis developed by Sigoillot, F.D., et al. (25).
3. RESULTS
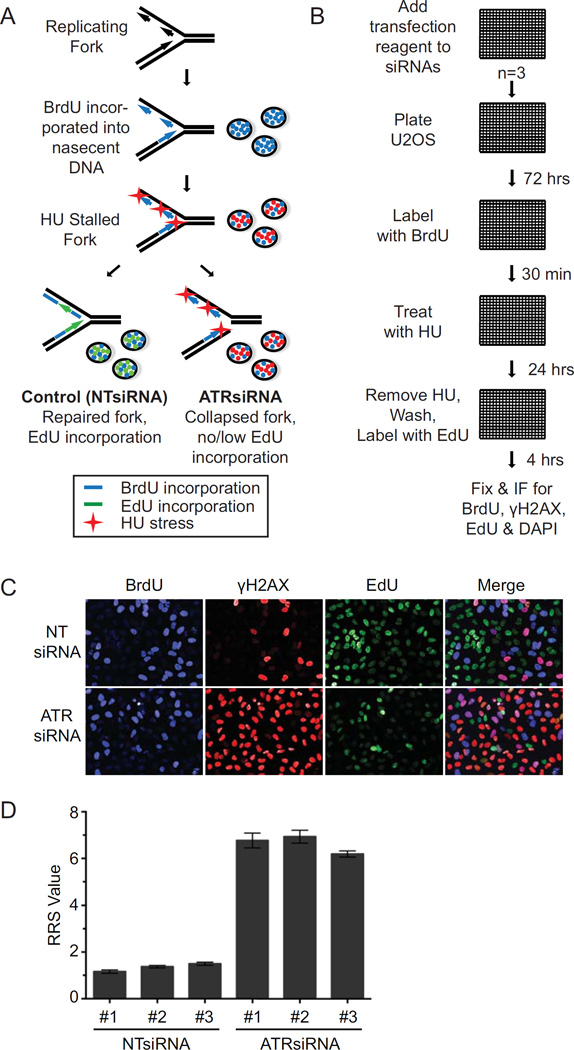
Our goal was to identify genes that function in the replication stress response using an unbiased siRNA screen (Fig 1A). Following siRNA transfection, effects on DNA synthesis were measured by incorporation of the thymidine analog BrdU. Following removal of BrdU, hydroxyurea (HU) (a ribonucleotide reductase inhibitor that diminishes deoxyribonucleotides availability for DNA synthesis) was added to cause replication stress (1). HU was subsequently removed and EdU added to monitor recovery of DNA synthesis from the replication stress challenge. Phosphorylation of histone H2AX (γH2AX) was also monitored as a second measure of how well the cells recovered from the replication stress challenge. In cells transfected with non-targeting control siRNA, fork damage will be repaired and replication resumed. This recovery is monitored by the disappearance of γH2AX and incorporation of EdU. When fork damage cannot be repaired and/or replication restarted, EdU incorporation will remain low and γH2AX levels high. We used ATR siRNA as a positive control for the screen.
Figure 1.
A whole genome siRNA screen for replication stress response genes. (A) Overview of the screening strategy. (B) U2OS cells were added to 384-well plates containing siRNA and Dharmafect transfection reagent. Seventy-two hours later, BrdU was incorporated for 30 minutes, then removed and cells were treated for 24 hours with 2mM HU. HU was removed and cells were labelled with 10µM EdU for 4 hours before fixing and performing immunofluorescence and automated imaging for BrdU, γH2AX, EdU, and DAPI. (C) Representative images of NT (non-targeting) and ATR siRNA controls showing BrdU and EdU incorporation and γH2AX intensity levels. (D) The mean values of NT and ATR control siRNAs from all plates for each replicate are depicted. Error bars represent standard error of the mean (SEM).
U2OS cells were transfected utilizing a whole genome siRNA library targeting 18,055 genes (four siRNAs per gene per well) in 384-well plates (Fig 1B). Each plate also contained non-targeting, ATR, and cell death siRNAs to serve as negative, positive, and transfection efficiency controls, respectively. After BrdU labelling, HU treatment, and EdU labelling, the cells were fixed and subjected to immunofluorescence imaging to detect BrdU, γH2AX, and EdU.
Nuclei were identified utilizing the DAPI signal and the intensity values of BrdU, γH2AX, and EdU for each nucleus determined via automated image analysis. As expected, efficient fork repair and restart resulting in high levels of EdU incorporation and low levels of γH2AX was observed in non-targeting control cells (Fig 1C). Conversely, ATR knockdown resulted in the appearance of many cells with low levels of EdU incorporation and high levels of γH2AX (Fig 1C). ATR knockdown did not significantly affect the incorporation of BrdU prior to the HU challenge.
The percentage of BrdU positive cells in each transfected cell population was determined to examine the effect of the loss of each gene on unperturbed replication (Suppl Table 1). The knockdown of most genes had no effect on the percentage of cells incorporating BrdU. The average percentage was 23.9 with a standard deviation of 6.6 percent (Suppl Fig 1A). Knockdown of one hundred ninety-two genes resulted in BrdU incorporation lower than three standard deviations from the mean and forty-five genes caused BrdU incorporation greater than three standard deviations above the population mean (Suppl Table 1). Genes with low BrdU incorporation when knocked down were enriched for processes involved in translation initiation, elongation and termination, RNA splicing and processing, and protein targeting (Suppl Fig 1B). These genes were excluded from further analysis since they are required generally for cell division making an assessment of their function in the replication stress response difficult. Gene knockdowns resulting in high BrdU incorporation percentages were enriched for genes involved in the cell cycle, mitotic processes, and checkpoints (Suppl Fig 1C).
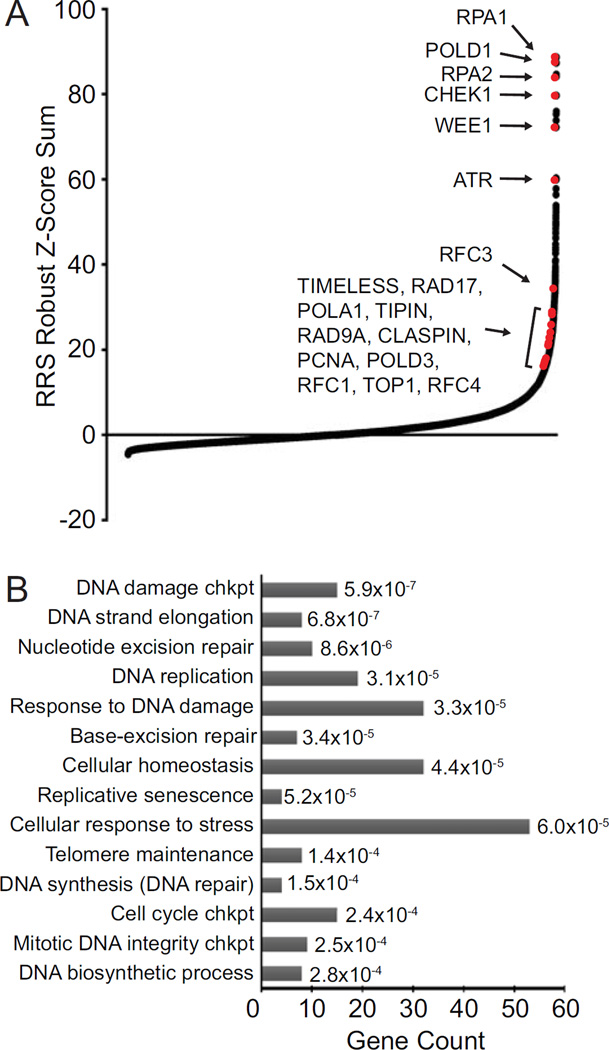
To determine siRNA effects on the replication stress response, a replication restart score (RRS) was calculated for each siRNA pool by first dividing the γH2AX intensity value by the EdU value for each nucleus. The RRS value was then determined by averaging the RRS for all nuclei within a sample. As anticipated, the ATR siRNA from all sample plates within each of the three replicates exhibited high mean RRS values (6.77, 6.93, and 6.19) in contrast to the non-targeting controls, which displayed low values (1.16, 1.37, and 1.49) (Fig 1D). The average RRS value for each gene was then used to calculate a robust Z-score on a plate-by-plate basis, and a robust Z-score sum using all three replicates of the screen for each gene (Fig 2A, Suppl Table 2). Five hundred twenty-four genes, or 2.9% of the total tested, had a Z-score sum of 15 or greater. ATR knockdown resulted in a Z-score sum of 59.94. Knockdown of multiple internal positive control genes caused high RRS scores including RPA1, RPA2, CHEK1, RFC1, RFC3, RFC4, TIMELESS, RAD17, POLA1, POLD1, POLD3, TIPIN, RAD9A, CLSPN, PCNA, and TOP1 (Fig 2A, Suppl Table 2). These genes have well-known functions in DNA replication and replication stress responses indicating that the screen was successful.
Figure 2.
The replication stress screen detects genes that function in DNA repair, DNA synthesis and stress responses. (A) Robust Z-score sum of the triplicate values of RRS for each gene are depicted. Several known replication and stress response genes, which served as positive controls, are indicated by red dots. (B) Gene ontology analysis of the top 400 scoring genes (ToppGene).
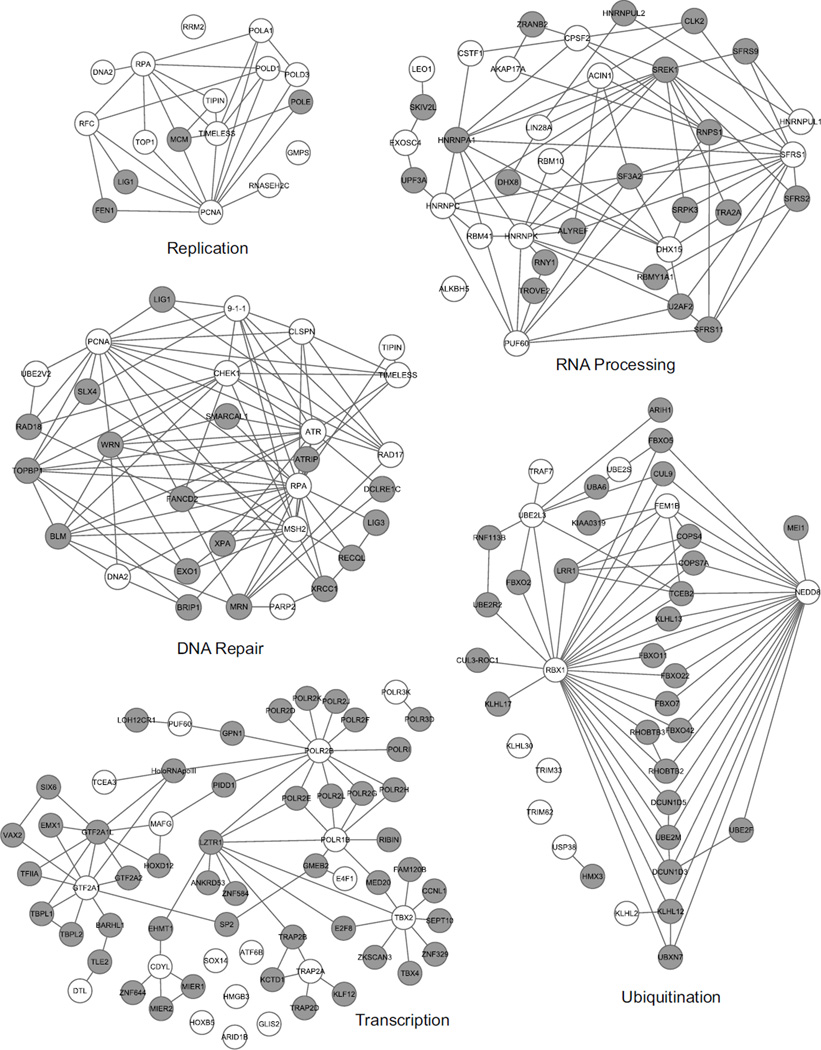
Gene ontology analysis for the top 400 genes (RRS Z-score sums of 17 or greater) indicated an enrichment in pathways linked to the replication stress response including: DNA damage checkpoint, DNA strand elongation, DNA replication, nucleotide and base excision repair, cellular response to DNA damage stimulus, cellular response to stress, DNA synthesis involved in DNA repair, cell cycle checkpoint, and DNA biosynthetic processes (Fig 2B and Fig. 3). Additionally, genes important for transcription, RNA processing, and ubiquitination were enriched among genes with the highest RRS scores (Fig 3, Suppl Table 2). Genes functioning in these pathways are critical for protein expression and function, and perturbations to these processes could impact the replication stress response indirectly or through impairment of regulatory mechanisms. RNA processing genes may also be detected due to increased conflicts between replication and transcriptional machineries such as R-loops (3).
Figure 3.
Network interaction maps illustrating genes identified by the screen in replication, DNA repair, RNA processing, transcription, and ubiquitination pathways. Genes identified from the screen are represented by white circles. Grey-shaded circles are genes known to interact either directly or indirectly with the identified genes. In some cases, genes encoding protein complexes like the MCM2-7 and Rad9-Hus1-Rad1 complex are grouped. (Ingenuity IPA and Cytoscape were utilized to generate interaction maps.)
To narrow the list of genes for validation and further study, high priority genes were identified to design a small custom library. First, genes with siRNAs that resulted in extensive cell death or severe impairments in cell division based on the BrdU analysis were removed from further consideration. The screen results were then compared to other ongoing replication screens in our laboratory as well as published screens investigating genes important for DNA damage responses and genomic integrity (12–23). The genes chosen for the custom library included the 200 with the highest RRS robust Z-score sums and genes detected in other screens. A few genes with known functions in replication and replication stress were included as internal positive controls but most were excluded. The custom library consisted of four individual siRNAs per gene and non-targeting, ATR, and cell death control siRNAs were included as controls.
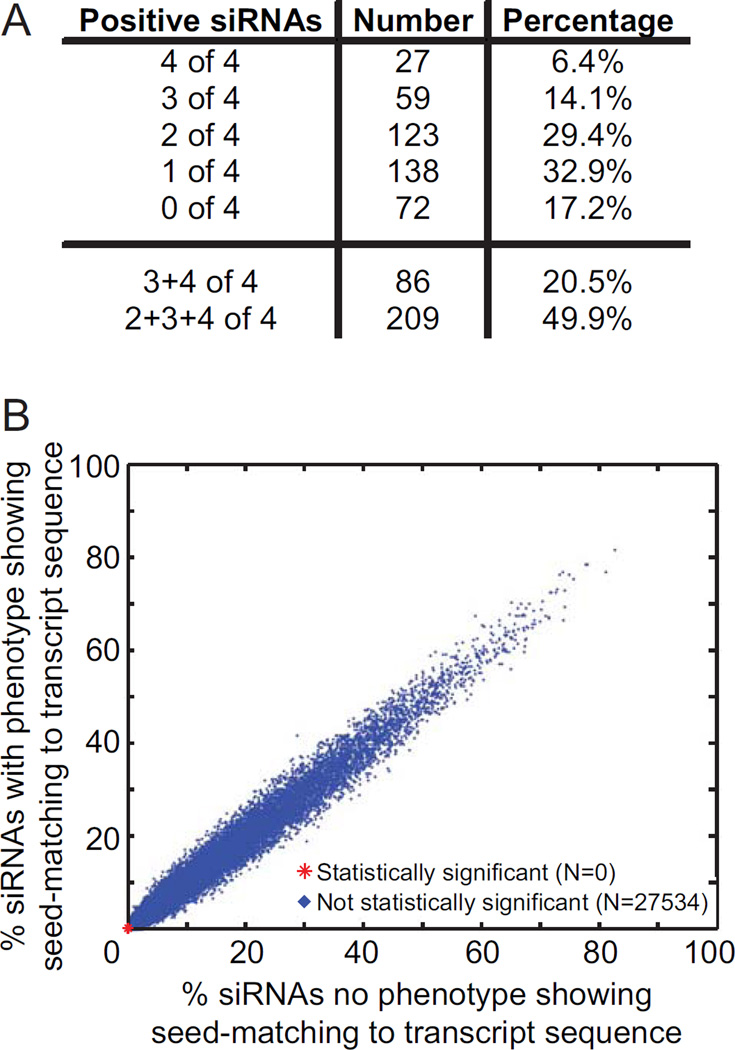
A validation screen was carried out using the same assay protocol as the whole genome screen. The data were examined to determine how many of the four individual siRNAs exhibited an increased RRS phenotype as observed in the pooled siRNA primary screen. The data was analysed using the Wilcoxon rank sum test to identify siRNAs that caused RRS values significantly different from the non-targeting siRNA control. Genes for which 2, 3, or 4 siRNAs yielded a false-discovery rate adjusted p-value less than or equal to 0.01 were considered to validate. A total of 49.9 percent of the genes analysed validated, with 6.4, 14.1, and 29.4 percent of the analysed genes validating with four, three, and two siRNAs, respectively (Fig 4A, Suppl Tables 3 & 4). All genes with known functions in DNA replication and repair that were included in the analysis as internal positive controls validated via this method. Additionally, genes known to function in cell cycle processes, stress response, chromosome organization, telomere maintenance, and cellular homeostasis also validated.
Figure 4.
Validation of the replication stress response screen to eliminate off-target effects. (A) Validation screen summary. The table indicates the number of siRNAs of 4 total per gene that exhibited a similar phenotype to the original result in the whole genome screen. The total gene number for each category and the overall percentage of validated genes is depicted. (B) GESS analysis of all siRNAs used in the screen to identify miRNA-like off-target effects.
To confirm that the siRNAs used in the validation screen did not result in off-target effects, we utilized the GESS analysis designed by Sigoillot, et al. (25). This analysis examines siRNA sequences to determine whether they also target miRNA seed sequences within the 3’- UTR region of genes that could result in off-target effects. The analysis did not detect significant off-target miRNA-like effects (Fig 4B), providing further confidence in our dataset.
To further assess the validated genes and determine which are important for cell growth and survival in stressed conditions, drug sensitivity screens were performed using HU, ATR inhibitor, or CHK inhibitor (Fig 5A, Suppl Fig 2). Increased sensitivity to HU would be expected for knockdown of replication stress response genes. Sensitivity to ATR or CHK inhibitors indicates the gene may function in the ATR and CHK1 pathway or its knockdown generates cellular conditions that require these pathways for survival (26). Sensitivity to additional drugs that promote replication stress – gemcitabine, a nucleoside analog that halts DNA synthesis and also inactivates ribonucleotide reductase; camptothecin, a topoisomerase inhibitor that causes DNA-TopoI adducts that interfere with replication; and Olaparib, a PARP inhibitor that traps PARP on DNA and blocks replication – were also examined (Suppl Figs 3 & 4). siRNAs causing cell viability at least 15 percent lower than non-targeting controls combined with an FDR-adjusted p-value of 0.01 or less were considered to cause sensitivity to drug treatment. At least two of the four siRNAs per gene were required to meet this threshold to consider a gene a hit. The complete dataset is presented in Suppl Table 5.
Figure 5.
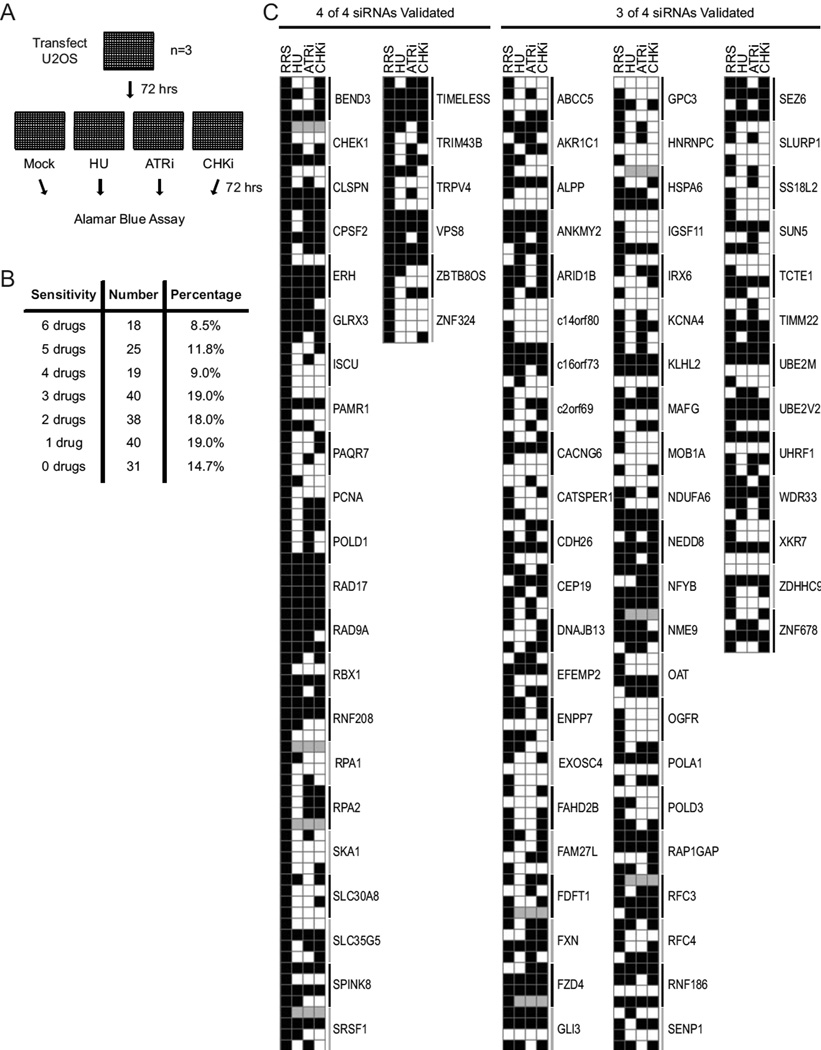
Knockdown of the identified replication stress response genes causes hypersensitivity to replication stress and ATR pathway inhibitors. (A) Flow-chart of the sensitivity screen assay. U2OS cells were transfected with siRNAs in 384-well plates. Seventy-two hours post-transfection cells were split 1:4 and were left untreated (mock) or treated with 0.2mM HU, 0.1µM ATR inhibitor, or 0.05µM CHK inhibitor for 72 hours before assaying for cell viability. (B) The number and percentage of genes required for resistance to drug treatments is indicated. Data from supplemental figures 2, 3, and 4, which include gemcitabine, camptothecin, and PARP inhibitor treatments, are included in the total count. (C) Results from validation and drug sensitivity screens are presented with each of the four siRNAs per gene as a row. The genes are grouped based on how many siRNAs validated in the RRS secondary screen (black squares in first column). Black squares in the remaining columns depict siRNAs causing sensitivity to the indicated drugs. Grey squares: not determined.
Of the 211 genes that validated from the original RRS screen, knockdown of 85.3 percent caused hyper-sensitivity to at least one drug, 66.4 percent caused sensitivity to at least two drugs, and 48.3 percent caused sensitivity to three or more drug treatments (Fig 5, Suppl Figs 2, 3 & 4). ATR siRNA positive controls exhibited sensitivity to all drugs tested. Knockdown of known components of the replication stress response including CLSPN, RAD17, RAD9A, TIPIN, TIMELESS, RPA2, and CHEK1 caused hypersensitivity to at least two drugs (Fig 5C, Suppl Figs 2, 3 & 4). Drug sensitivity of these known replication stress response genes gives confidence that other genes displaying sensitivity within the dataset could function in DNA replication, repair and the replication stress response.
As expected, hydroxyurea sensitivity was a common phenotype associated with the genes identified in the primary RRS screen. However, this was not a universal affect possibly because the sensitivity and RRS screens utilized different HU concentrations and different durations of treatment. There is also a strong correlation between siRNAs that yielded phenotypes in the RRS screen and those that yield hypersensitivity to drug treatments.
4. DISCUSSION
We designed and implemented a whole genome siRNA screen assay utilizing thymidine analog incorporation and HU treatment to identify replication stress response genes. We identified internal positive controls including ATR, RPA, CHEK1 and RAD9A providing confidence that the screen performed as expected. Furthermore, analysis of the top 400 genes from the primary screen revealed enrichment of genes that function in cell cycle checkpoints, DNA replication, DNA repair, and cellular responses to stress. Other pathways that were enriched included transcription, RNA processing, and ubiquitination. The genes in these pathways regulate many cellular functions and can have pleiotropic effects. As examples, RBX1, which interacts with cullins and plays an essential role in ubiquitin polymerization to target proteins for ubiquitin-mediated destruction, and NEDD8, which is necessary for the NEDDylation of cullins to facilitate ubiquitin conjugation, were identified. The absence of either RBX1 or NEDD8 affects the ubiquitination of many other proteins ultimately interfering with proper control of protein expression, cell cycle control and replication stress responses.
To ensure our dataset removes off-target effects, we validated the primary screen with individual siRNAs and a stringent cut-off for both statistical significance and magnitude of response. Several of the validated genes have previously been identified in other published genome stability and DNA repair screens. For example, CHERP, CLIC3, CRYAA, ERH, FDFT1, GLRX, HSPA6, LMNB1, MGST3, MUC7, NCBP1, OAT, ORJ2J, RAP2C, RNF26, UBL3, and ZNF324 were identified in a screen for genes whose knockdown causes an increase in spontaneous DNA damage (12). A proteomic screen for ATM/ATR substrates identified FLJ20516, NFRKB, OGFR, PHF3, and RBM10 (18), and purification of proteins on nascent chromatin identified AHCY, HLA-DRB5, HSPA6, and LMNB1 (27). Detection of these genes in other screens examining genome stability, damage repair pathways, and replication further supports the likelihood that they function in the replication stress response.
Analysis of validated genes to determine how their loss of function affects viability in response to HU, ATR inhibitor, CHK inhibitor, gemcitabine, camptothecin, and PARP inhibitor found that 85 percent caused hyper-sensitivity to at least one drug and over 66 percent caused hyper-sensitivity to multiple drug treatments. Cellular sensitivity to HU, gemcitabine, and/or camptothecin would be predicted for genes needed for completion of DNA synthesis in the context of replication stress. PARP inhibitors trap PARP on DNA, block replication, and cause an increased requirement for recombination to repair the damage. Thus, PARP inhibitor sensitivity may indicate genes involved in replication-associated recombination pathways. Sensitivity to ATR or CHK inhibitors suggests the gene is associated with inactivation of DNA repair pathways or even partial inactivation of the ATR pathway itself since partial inactivation of the pathway generates sensitivity to further inactivation via inhibitor action (26). Knockdown of some genes caused an inability to recover from HU, but did not cause hyper-sensitivity to HU in the viability assay. This may be explained by differences in drug concentration and the duration of treatment between the two assays. These datasets provide further validation that the gene list is enriched in replication stress response activities and will help direct further investigation into the function of proteins encoded by these genes.
As one example, knockdown of the gene ERH (enhancer of rudimentary homolog) caused sensitivity to HU, gemcitabine, camptothecin, and ATR, CHK, and PARP inhibitors. Further studies confirmed ERH indeed effects replication via a role in mRNA splicing of ATR and a subset of other genes important for processing replication stress and DNA damage (28). Several other genes that when knocked down exhibited sensitivity to multiple drugs may also be interesting candidates for further study. RNF208, of which little is known regarding its function, displayed sensitivity to HU and ATR and CHK inhibitors when knocked down. c16orf73, which is important for meiotic crossovers and contains an OB-fold domain that binds ssDNA (29–31), warrants further examination. Knockdown of SENP1 led to HU, gemcitabine, camptothecin, and CHK and PARP inhibitor sensitivity. SENP1 functions in sumoylation processing (32–35), and therefore could possibly affect replication stress proteins via alterations in sumoylation.
In summary, our study provides an overview of genes that impact the replication stress response. Many may function indirectly in signalling, gene expression, metabolism or other cellular processes that impinge on replication and repair. Others are likely to encode proteins that directly function at replication forks. In either case, the data will provide a useful resource for investigators interested in cell division, replication, and replication stress responses.
Supplementary Material
Highlights.
A three parameter RNAi screen identified new replication stress response genes
Candidate replication stress response genes were validated in secondary screens
The dataset provides a resource for understanding the replication stress response
ACKNOWLEDGEMENTS
The Vanderbilt Institute for Integrative Biosystems Research and Education core facility was utilized for high content imaging. This work was supported by the National Institutes of Health [F32GM103254 to G.K., T32ES007028 to J.W.L., and R01CA102729 to D.C.] and Susan G. Komen [PDF14302198 to K.N.M]. Acquisition of the Opera automated microscope was supported by 1S10RR027485.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 2.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 3.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 6.Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell. Biol. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 10.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 11.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 12.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovejoy CA, Xu X, Bansbach CE, Glick GG, Zhao R, Ye F, Sirbu BM, Titus LC, Shyr Y, Cortez D. Functional genomic screens identify CINP as a genome maintenance protein. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19304–19309. doi: 10.1073/pnas.0909345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu DS, Zhao R, Hsu EL, Cayer J, Ye F, Guo Y, Shyr Y, Cortez D. Cyclin-dependent kinase 9-cyclin K functions in the replication stress response. EMBO Rep. 2010;11:876–882. doi: 10.1038/embor.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotta-Ramusino C, McDonald ER, 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 19.Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J. Biol. Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 20.Moudry P, Lukas C, Macurek L, Neumann B, Heriche JK, Pepperkok R, Ellenberg J, Hodny Z, Lukas J, Bartek J. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ. 2012;19:798–807. doi: 10.1038/cdd.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, Schlabach M, Gygi SP, Elledge SJ, Harper JW. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Molecular cell. 2010;40:645–657. doi: 10.1016/j.molcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci. Signal. 2010;3:rs3. doi: 10.1126/scisignal.2001034. [DOI] [PubMed] [Google Scholar]
- 23.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini YaHY. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 25.Sigoillot FD, Lyman S, Huckins JF, Adamson B, Chung E, Quattrochi B, King RW. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014;74:2835–2845. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kavanaugh G, Zhao R, Guo Y, Mohni KN, Glick G, Lacy ME, Hutson MS, Ascano M, Cortez D. Enhancer of Rudimentary Homolog affects the replication stress response through regulation of RNA processing. Mol. Cell. Biol. 2015 doi: 10.1128/MCB.01276-14. pii: MCB.01276-14. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joyce EF, Tanneti SN, McKim KS. Drosophila hold'em is required for a subset of meiotic crossovers and interacts with the dna repair endonuclease complex subunits MEI-9 and ERCC1. Genetics. 2009;181:335–340. doi: 10.1534/genetics.108.093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo M, Yang F, Leu NA, Landaiche J, Handel MA, Benavente R, La Salle S, Wang PJ. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat. Commun. 2013;4:2788. doi: 10.1038/ncomms3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souquet B, Abby E, Herve R, Finsterbusch F, Tourpin S, Le Bouffant R, Duquenne C, Messiaen S, Martini E, Bernardino-Sgherri J, et al. MEIOB targets single-strand DNA and is necessary for meiotic recombination. PLoS Genet. 2013;9:e1003784. doi: 10.1371/journal.pgen.1003784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen LN, Dong C, Liu H, Naismith JH, Hay RT. The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem. J. 2006;397:279–288. doi: 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JH, Lee SW, Yang SW, Yoo HM, Park JM, Seong MW, Ka SH, Oh KH, Jeon YJ, Chung CH. Modification of DBC1 by SUMO2/3 is crucial for p53-mediated apoptosis in response to DNA damage. Nat. Commun. 2014;5:5483. doi: 10.1038/ncomms6483. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Wang M, Ao X, Chang AK, Yang C, Zhao F, Bi H, Liu Y, Xiao L, Wu H. CLOCK is a substrate of SUMO and sumoylation of CLOCK upregulates the transcriptional activity of estrogen receptor-alpha. Oncogene. 2013;32:4883–4891. doi: 10.1038/onc.2012.518. [DOI] [PubMed] [Google Scholar]
- 35.Cheng J, Wang D, Wang Z, Yeh ET. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 2004;24:6021–6028. doi: 10.1128/MCB.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.