Abstract
Humans carry two copies of Survival Motor Neuron gene: SMN1 and SMN2. Loss of SMN1 coupled with skipping of SMN2 exon 7 causes spinal muscular atrophy (SMA), a leading genetic disease associated with infant mortality. Our discovery of intronic splicing silencer N1 (ISS-N1) is a promising target, currently in phase 3 clinical trial, for an antisense-oligonucleotide-mediated splicing correction in SMA. We have recently shown that the first residue of ISS-N1 is locked in a unique RNA structure that we term ISTL1 (Internal Stem Through Long-distance interaction-1). Complementary strands of ISTL1 are separated from each other by 279 nucleotides. Using site-specific mutations and chemical structure probing we confirmed the formation and functional significance of ISTL1. Located in the middle of intron 7, the 3′ strand of ISTL1 falls within an inhibitory region that we term ISS-N2. We demonstrate that an antisense-oligonucleotide-mediated sequestration of ISS-N2 fully corrects SMN2 exon 7 splicing and restores high levels of SMN in SMA patient cells. These results underscore the therapeutic potential of the regulatory information present in a secondary and high-order RNA structure of a human intron.
Introduction
Formation of RNA structure is essential for many steps in controlled gene expression, including regulation of alternative splicing.1–3 There is a growing acceptance of the role terminal stem-loop (TSL) structures play in modulating the accessibility of splice sites (ss).2 Advancements in computational algorithms have allowed predictions of complex RNA structures. Often, multiple secondary structures could be predicted for the same transcript and the probability of structural variants increases with the increase in the size of the sequence. However, extracting the significance of a particular structure among multiple alternatives remains a daunting task. Consequently, it is still a mystery how the splicing machinery is guided by certain RNA structures to remove specific intronic sequences. In the absence of a simple method of structure determination in vivo, compensatory mutations combined with in vitro structure probing (chemical and/or enzymatic) provide evidence in support of an RNA structure that functions in splicing. Overall the process of structure validation remains an arduous endeavor with very limited studies available on probed RNA structures of large human pre-mRNAs.
About half of all human genetic disorders are caused by mutations that alter pre-mRNA splicing.4 Growing evidence suggests that significant regulatory information is trapped in RNA structures of transcriptomes.5,6 Of note, ~40% of the human genome are intronic sequences.7 Hence, there is a need for developing functional assays that uncover the role of critical intronic RNA structures in regulation of pre-mRNA splicing. Here we describe a unique RNA structure formed by long-distance interactions (LDI) between intronic sequences within a critical gene associated with spinal muscular atrophy (SMA). SMA is the leading genetic disease of children and infants. Detailed aspects not covered in this report, such as SMA pathogenesis, demography of disease, mouse models of SMA and therapeutic approaches, have been described in recent reviews.8–14
Molecular basis of SMA
Humans have two nearly identical copies of the Survival Motor Neuron (SMN) gene: SMN1 and SMN2.15 Two SMN genes code for identical proteins; however, due to skipping of exon 7, SMN2 predominantly generates a shorter transcript, which is translated into a truncated, unstable protein.16,17 The inability of SMN2 to compensate for the loss of SMN1 results in SMA, a debilitating childhood disease.18 SMN is an essential housekeeping protein with multiple functions that include snRNPs biogenesis, transcription, translation, signal transduction, stress granule formation and macromolecular trafficking. Several recent reviews describe SMN functions in detail.10,11,19–21 Here we focus on the evolving mechanism of splicing regulation of SMN2 exon 7, since strategies aimed at the postnatal restoration of exon 7 inclusion in SMN2 have shown promise for SMA therapy.
SMN2 mutations associated with exon 7 skipping
A C-to-T mutation at the 6th position (C6U in transcript) of exon 7 and an A-to-G substitution at the 100th position (A100G) of intron 7 contribute towards skipping of SMN2 exon 7 (Fig. 1).22,23 To explain the inhibitory effects of C6U, various mechanisms including loss of an enhancer associated with SF2/ASF, gain of a silencer associated with hnRNP A1 and strengthening of a stem-loop structure (TSL1) at the 3′ ss of exon 7 have been proposed (Fig. 1).23–29 It has also been suggested that C6U creates an extended inhibitory context (Exinct) that encompasses most of the TSL1.50 Consistently, substitutions within Exinct/TSL1 have been found to restore SMN2 exon 7 inclusion.
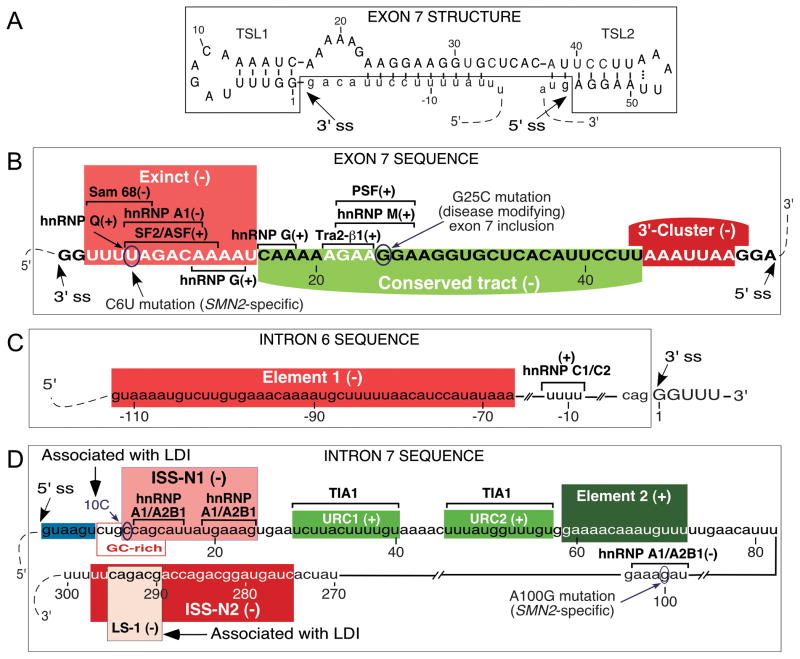
Figure 1.
Diagrammatic representation of cis-elements for SMN exon 7 splicing. (A) Secondary structure of SMN2 of exon 7 based on enzymatic probing.24 Numbering starts from the beginning of exon 7. (B) Diagrammatic representation of cis-elements within exon 7. Numbering starts from the beginning of exon 7. Positive and negative cis-elements/transacting factors are indicated by (+) and (−), respectively. Exinct (Extended inhibitory context), Conserved tract and 3′-Cluster were identified by in vivo selection.25 Binding sites for SF2/ASF, hnRNP A1, hnRNP A2/B1, Sam 68, hnRNP Q, Tra2-β1, hnRNP G, hnRNP M and PSF described elsewhere.26–36 G-to-C mutation at position 25 (G25C) in SMN2 exon 7 has been shown to modify the severity of SMA.37–39 (C) Diagrammatic representation of cis-elements within intron 6. Nucleotides of intron 6 and exon 7 are shown in lower case and capital letters, respectively. Numbering starts from the beginning of exon 7. Location of Element 1 is highlighted in red.40 PTB and FUSE-BP interact with Element 1.41 A binding site of the stimulatory hnRNP C1/C2 is indicated.42 (D) Diagrammatic representation of cis-elements within intron 7. Numbering starts from the beginning of intron 7. Positive and negative cis-elements are indicated by (+) and (−), respectively. The 5′ ss of exon 7 is highlighted in blue. ISS-N1, the overlapping GC-rich sequence as well as 10C and LS-1 that are engaged in LDI with each other, all contribute to skipping of exon 7.43–46 ISS-N1 harbors two putative hnRNP A1/A2B1 binding sites.47 An A100G substitution creates a binding site for hnRNP A1.23 Element 2 and U-rich clusters (URC1 and URC2) are positive cis-elements.48, 49 Stimulatory factor TIA1 interacts through URC1 and URC2.49 ISS-N2 is a structure-associated inhibitory element located in the 3′ half of intron 7.46
In vivo selection of the entire exon
To determine the impact of every single exonic residue on splicing, we performed an in vivo selection of the entire exon 7.25 Briefly, SMN1 minigene containing partially randomized exon 7 was transfected into human cervical carcinoma (C33a) cells. The purpose of partial randomization was to maintain the wild type characteristics of the exon while probing the position-specific significance of every residue within the 54-nt long exon 7.51 About 20 h posttransfection total RNA was isolated and reverse transcribed using oligo dT primer. Thereafter, exon 7-included transcripts were amplified using engineered primers encompassing restriction sites for re-cloning into SMN1 minigene splicing cassette. The process was repeated four times to enrich exon 7 sequences that favored inclusion of exon 7. About sixty unique sequences of exon 7 from the final selected pool were analyzed for regulatory motifs. Exonic positions where wild-type residues were preserved were considered as positive, whereas, exonic positions where wild-type residues were substituted by non-wild type residues were considered as negative. Motifs were defined based on the clustering of the positive and the negative positions. Findings of in vivo selection validated the presence of Exinct and revealed another inhibitory region (3′-Cluster) at the end of exon 7 (Fig. 1B).25 The middle portion of exon 7 was found to harbor a positive regulatory region termed Conserved tract (Fig. 1B). The above findings of in vivo selection were independently validated by an antisense-oligonucleotide (ASO)-based approach.52
The last position of exon 7 is an A residue. However, a non-wild type G residue (54G) was overwhelmingly selected at this position.25 Our subsequent experiments showed that 54G was able to fully restore SMN2 exon 7 inclusion even in the absence of several positive regulatory elements that were originally thought to be critical for exon 7 splicing. When we reported the significance of 54G, most studies focused on the 3′ ss of exon 7 due to its (3′ ss) close proximity to C6U mutation, which at the time was thought to be the primary cause of exon 7 skipping.53 The results of in vivo selection brought a new perspective to the field (of exon 7 splicing regulation) by demonstrating an intrinsic problem with the 5′ ss.25 This prompted us to further characterize the 5′ ss that is defined by the linear RNA sequence as well as RNA secondary structure at the junction of exon 7 and intron 7.
Weak 5′ ss as a limiting factor for exon 7 splicing
The strong stimulatory effect of 54G on SMN2 exon 7 splicing can be explained by improved recruitment of U1 snRNP to the 5′ ss due to the increase in the number of base pairs formed between U1 snRNA and the 5′ ss. It is also possible that the stimulatory effect of 54G is due to the disruption of a predicted stem-loop structure (TSL2) that sequesters the 5′ ss of exon 7.24 Compensatory mutations and enzymatic structure probing validated the formation of TSL2. Also, mutations that disrupted TSL2, restored SMN2 exon 7 inclusion, confirming that the poor accessibility of the 5′ ss contributes to exon 7 skipping. Further validating that the 5′ ss of exon 7 is poorly defined, a mutant U1 snRNA that increased the base pairing with the wild type 5′ ss of exon 7 restored SMN2 exon 7 inclusion.24 Presence of the intronic splicing silencer N1 (ISS-N1) immediately downstream of the 5′ ss of exon 7 provided an additional basis for the weak 5′ ss of this exon (Fig. 1D).43 Of note, the 15-nt long ISS-N1 is a portable cis-element predicted to harbor two hnRNP A1 motifs.43,47 Deletion or an ASO-mediated sequestration of ISS-N1 promoted SMN2 exon 7 inclusion. ISS-N1 partially overlaps with an 8-nt long GC-rich sequence (GCRS), sequestration of which by an 8-mer ASO also promoted SMN2 exon 7 inclusion.44,54 Notably, ISS-N1 has emerged as the most tested antisense target for splicing correction in a human genetic disease.11 Currently, an antisense drug (ISIS-SMNRx) based on ISS-N1 target is undergoing phase 3 clinical trial by ISIS Pharmaceuticals.55
Role of transacting factors on SMN2 exon 7 splicing
Multiple factors, including SF2/ASF, hnRNP A1, Tra2-β1, hnRNP G, hnRNP Q, Sam68, and TIA1 have been implicated in regulation of SMN2 exon 7 splicing (Fig. 1). Most of the reported factors act through binding to exon 7. Tra2-β1 that directly interacts with exon 7 and recruits SRp30c, hnRNP G and TDP-43 to exon 7 was initially thought to be critical for exon 7 inclusion.32,56–58 However, latest observations in a Tra2-β1 knockout mouse support the dispensable nature of Tra2-β1 for SMN exon 7 splicing.59 Recently, Welander distal myopathy (WDM) patients carrying TIA1 mutations have been found to exhibit an enhanced rate of SMN exon 7 skipping.60 TIA1 is a glutamine (Q)-rich RNA-binding protein (QRDP) that is known to recruit U1 snRNP to the 5′ ss of exons.61,62 We have previously shown that TIA1 interacts with two U-rich clusters (URC1 and URC2) within intronic sequences downstream of ISS-N1 and that overexpression of TIA1 restores SMN2 exon 7 inclusion (Fig. 1D).49 PSF, another QRDP has been recently shown to promote SMN2 exon 7 inclusion by binding to an exonic sequence that overlaps Tra2-β1 binding site.36 Out of four binding sites of hnRNP A1, three are located within intron 7 (Fig. 1D). While first two intronic binding sites of hnRNP A1 are located within ISS-N1, the third one is associated with SMN2-specific mutation at the 100th position of intron 7 (Fig. 1D).23,47 Of note, the role of many proteins shown to affect SMN2 exon 7 splicing in cell-based assays have not yet been validated in knockout mouse models and/or in patients.
Discovery of a unique LDI
For simplicity we define a LDI as any RNA:RNA interaction in which two distantly located sequences (or residues) are brought together by canonical or non-canonical base pairing. In certain situations, LDIs could be formed by residues separated form each other by thousands of nucleotides. LDI prediction as well as validation of their functional significance in pre-mRNA splicing is quite challenging. We have recently reported the critical role of a unique RNA structure formed by LDIs within SMN2 intron 7.46 One of the defining aspects of our discovery is the ASO-based approach that uncovered the critical role of a cytosine residue at the 10th intronic position (10C) in the formation of this inhibitory RNA structure.45 The discovery of a 10C-mediated LDI is significant since it offers several antisense targets as potential therapies for SMA.
Functional assay leading to the discovery of LDI
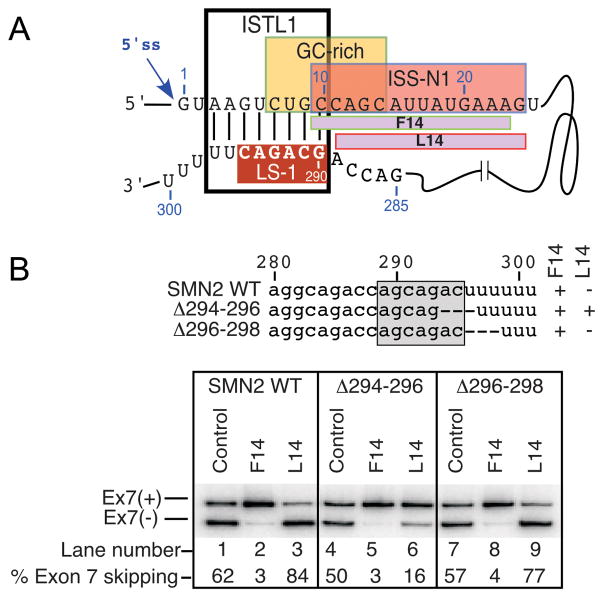
While 10C is the first position of ISS-N1, it is also a part of GCRS that partially overlaps with ISS-N1.44 The structure-associated role of 10C was serendipitously discovered in an antisense microwalk in which two 14-nt long ASOs (F14 and L14) produced opposite effects on SMN2 exon 7 splicing (Fig. 2).45 While F14 promoted SMN2 exon 7 inclusion by sequestering the first 14 residues of 15-nt long ISS-N1, L14 promoted SMN2 exon 7 skipping by sequestering the last 14 residues of ISS-N1. Substitutions or deletion of 10C or deletion of a deep intronic sequence in the 3′ half of intron 7 abrogated the inhibitory effect of L14.45 Therefore, we attributed the inhibitory effect of L14 to a LDI in which 10C interacts with downstream intronic sequences. To narrow down the exact site of the LDI, we made overlapping deletions throughout the intron 7 in an SMN2 minigene construct and compared the splicing pattern of these mutants in the presence and absence of L14. We identified a 6-nt long motif (GCAGAC) spanning the region from 290th to 295th positions of intron 7 as the interacting partner of 10C (Fig. 2).46 We named this motif the LDI Site 1 or LS-1. The entire LS-1 sequence was predicted to be engaged in the formation of an internal stem (RNA:RNA duplex) made by LDI. We termed this structure as an internal stem through LDI 1 or ISTL1 (Fig. 2). Confirming the association of ISTL1 with the inhibitory effect of L14, mutations that disrupted ISTL1 fully eliminated the negative effect of L14 on exon 7 splicing. Interestingly, L14 does not interact with any of the residues that constitute ISTL1, since its annealing site is located downstream of ISTL1, and yet it (L14) exerts its negative effect on exon 7 splicing through ISTL1. These findings underscore an important but rarely observed phenomenon that the effect of an ASO could be executed indirectly through structures formed in the vicinity of the ASO annealing site.
Figure 2.
Intronic sequences involved in LDI. (A) Diagrammatic representation of F14 and L14 binding sites within ISS-N1. Relative positioning of ISS-N1, GCRS, LS-1 and ISTL1 are shown. (B) Effect of F14 and L14 on SMN2 exon 7 splicing. Experiments were done using SMN2 minigenes.45 Sites of intronic deletions are shown by dotted lines (top panel). Bottom panel shows loss of inhibitory effect of L14 due to a 3-nt deletion that abrogates ISTL1.
Structure probing validated the formation of ISTL1
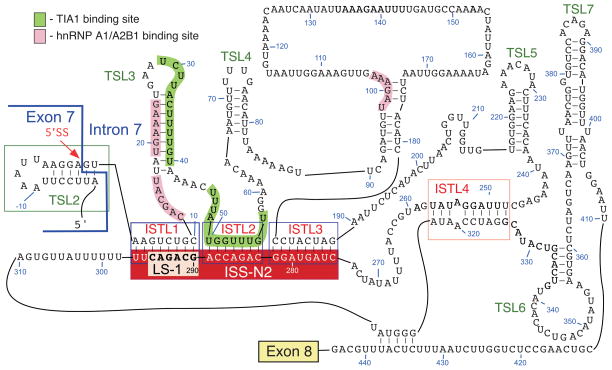
The formation of ISTL1 implicated the deep intronic sequence in influencing the decisions made at the 5′ ss of SMN2 exon 7. To confirm that ISTL1 is indeed formed, we probed the structure of the entire SMN2 intron 7 employing Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) method.46 Among available methods of in vitro structure probing, SHAPE is a relatively unbiased technique that allows simultaneous interrogation of all residues within an RNA of interest.63 The structure probed by SHAPE confirmed the formation of ISTL1. ISTL2, ISTL3 and ISTL4 emerged as additional structures formed by LDIs (Fig. 3). The results of structure probing placed the first hnRNP A1 binding site associated with ISS-N1 within an internal loop between ISTL1 and TSL3 (Fig. 3).46 The second hnRNP A1 binding site associated with ISS-N1 constituted the 5′ stem of TSL3 (Fig. 3). The hnRNP A1 motif at the 100th position of intron 7 was engaged in short internal stem formation (Fig. 3). According to the SHAPE analysis, URC1 and URC2 are located in double-stranded regions (Fig. 3). This structural arrangement is likely to prevent recruitment of TIA1 to these sites. Our experimentally derived structure did not support some of the predictions made for several regions of intron 7, including an Element 2-associated TSL located downstream of URC2.48 The results of the SHAPE analysis validated the formation of TSL2, although the U residue at the second intronic position (2U) was found to be accessible (Fig. 3). The accessibility of 2U could be due to co-axial stacking of bases at the junction of TSL2 and ISTL1.
Figure 3.
Structure of SMN2 intron 7 deduced by chemical probing, using SHAPE. Positions of LS-1, ISTLs, TSLs and binding sites of hnRNP A1/A2B1 and TIA1 are shown. ISS-N2 is comprised of 3′ strands of ISTL1, ISTL2 and ISTL3.46 Numbering starts from the beginning of intron 7.
Mechanism of ISTL1-mediated splicing regulation
ISTL1 is an 8 base-pair duplex with 50% GC-rich content. The 5′ strand of ISTL1 is separated from the 3′ strand by 279 residues (from 11th to 289th positions of intron 7). The distal end of the duplex is closed by a GC base pair formed between 10C and 290G, whereas, the proximal end of the duplex is closed by AU base pair formed between 3A and 297U. The side-by-side positioning of TSL2 and ISTL1 fully sequesters the U1 snRNA-annealing site at the 5′ ss of exon 7 (Fig. 4). Hence, ASOs that disrupt ISTL1 are likely to induce recruitment of U1 snRNP and stimulate SMN2 exon 7 inclusion. Based on the findings of structure probing in the presence of L14 and F14, we conclude that ISTL1 is stabilized and destabilized by L14 and F14, respectively.46 Interestingly, a major reduction in size of intron 7 by deletions from 93rd to 281st positions and from 330th to 412th positions of intron 7 stimulated SMN2 exon 7 inclusion.46 As per structure prediction these deletions do not disrupt ISTL1; they preserve the inhibitory effect of L14. Similarly, breaking of ISTL2 by three nucleotide deletions stimulated SMN2 exon 7 inclusion; note that in the presence of these deletions ISTL1 structure is predicted to be maintained and L14 remains inhibitory.46 The above result of deletion mutations suggests that the overall structural context of intron 7 has a stabilizing effect on ISTL1. However, since the stimulatory effect of deletions could be due to the loss of negative elements and/or gain of positive elements, additional experiments will be required to assess the role of other structures on the stability of ISTL1. Our finding that L14 remains inhibitory in deletion mutants described above suggests that the stabilization of ISTL1 by L14 does not require most of the intronic structures. Our results also support that hnRNP A1/A2B1 and PTB proteins are not involved in the formation of ISTL1, since their depletion did not abrogate the inhibitory effect of L14.46 However, one cannot rule out the role of yet unknown proteins in ISTL1 formation/stabilization.
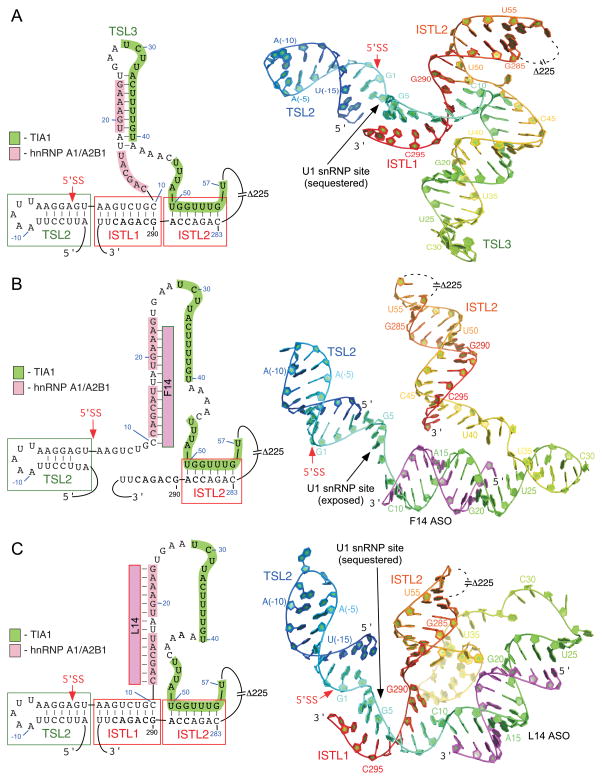
Figure 4.
The 3D structure models at the 5′ ss of exon 7 based on the SHAPE derived secondary structure of intron 7. The predicted 3D conformations and tertiary structures are modeled on chemical structure probing and not defined by experimental constraints.46 (A) The probed secondary structure (left panel) and the predicted 3D structure model (right panel) of the wild type 5′ ss of exon 7. 3D Model was predicted using the RNA assembly application within the Rosetta modeling suite.64 Numbering starts from the beginning of intron 7 at G1 with a rainbow color scheme ranging from blue at the 5′ end to red at the 3′ end. Relative positioning of TSL2 (residues −17 to 2), TSL3 (residues 17 to 41), ISTL1 (residues 3 to 10 and 290 to 297) and ISTL2 (residues 50 to 56 and 283 to 289) are shown. The TSL3 stem is extended by mismatched base pairing (residues 12 to 16 and 42 to 46), which is not defined by experimental data and may be transient. Model shows a sequestered 5′ ss that is not favorable for the recruitment of U1 snRNP. (B) The probed secondary structure (left panel) and the 3D structure model (right panel) of the 5′ ss of exon 7 in the presence of F14. Model was predicted similarly as described for the wild type context in section A. F14 is highlighted in magenta with the 5′ and 3′ ends indicated. The retained secondary structural elements are labeled, which includes TSL2 (residues −17 to 2) and ISTL2 (residues 50 to 56 and 283 to 289). The stem region of ISTL2 is extended by mismatch base pairing (residues 42 to 49 and 290 to 298) and an internal bulge (C291). Model shows an accessible 5′ ss that is favorable for the recruitment of U1 snRNP. (C) The probed secondary structure (left panel) and the 3D structure model (right panel) of the 5′ ss of exon 7 in the presence of L14. Model predictions and labeling details are the same as described in section B. Model shows a sequestered 5′ ss that is not favorable for the recruitment of U1 snRNP.
LDIs as targets for SMA therapy
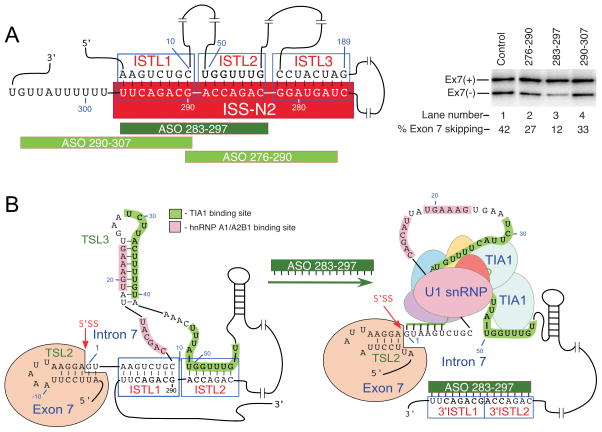
The 3′ strands of adjacent stems ISTL1, ISTL2 and ISTL3 are formed by the deep intronic sequence from positions 275 to position 297, we called this sequence ISS-N2 (Figs. 1 & 5). Located far away from the 5′ss, ISS-N2 emerged as a unique sequence engaged in formation of three adjacent structures. Underscoring the inhibitory role of ISS-N2, deletions within ISS-N2 stimulated SMN2 exon 7 inclusion.46 Therefore, we evaluated ISS-N2 as a potential target for an ASO-mediated splicing correction in SMA patient cells.
Figure 5.
ISS-N2 as a target for splicing correction in SMA. (A) Left panel shows diagrammatic representation of ISS-N2 target and ASOs annealing positions, whereas, right panel shows the effect of ASOs on SMN2 exon 7 splicing in SMA patient cells. Experimental details and results have been described elsewhere.46 (B) Proposed mechanism by which ASO 283-297 promotes exon 7 inclusion. ASO 283-297 disrupts ISTL1 and ISTL2, leading to binding of TIA1 and enhanced recruitment of U1 snRNP.
ISS-N2-targeting ASOs stimulate SMN2 exon 7 inclusion
We employed SMA-patient-derived GM03813 cell line to test whether blocking of ISS-N2 by ASOs will have a positive effect on SMN2 exon 7 splicing. GM03813 cells contain only SMN2 and have been widely used to test the efficacy of compounds in modulating exon 7 splicing. Our exploratory experiments were performed with propriety-free RNA ASOs containing phosphorothioate backbone and 2′-O-methyl modifications at every sugar residue. To minimize the off-target effect, we performed experiments at low nM ASO concentrations. Among eight ASOs tested, ASO 283-297 that sequestered the 3′ strands of ISTL1 and ISTL2 led to a substantial inclusion of SMN2 exon 7 (Fig. 5).46 Consistent with the increase in exon 7 inclusion, ASO 283-297 elevated the levels of SMN and SMN-interacting protein Gemin2. These results validated that a deep intronic sequence associated with an inhibitory structure formed by a LDI could serve as a potential target for SMA therapy.
Mechanism of ISS-N2-targeting ASO
Stimulation of SMN2 exon 7 splicing by ASO 283-297 could occur through a combinatorial effect leading to the enhanced recruitment of U1 snRNP at the 5′ ss of exon 7. Indeed, a recent report suggests a multitude of interactions at the 5′ ss of exon 7.65 Annealing of ASO 283-297 to its target disrupts ISTL1 and ISTL2 freeing the 5′ ss for interactions with U1 snRNP and rendering URC1 and URC2 accessible for binding by TIA1, which is known to recruit U1 snRNP to weak 5′ ss. Therefore, simultaneous disruption of ISTL1 and ISTL2 is likely to have additive or synergistic effect on the recruitment U1 snRNP to the 5′ ss of exon 7. Consistent with this hypothesis, ASO 290-307 that disrupted ISTL1 but not ISTL2 produced a lesser stimulatory effect on SMN2 exon 7 splicing (Fig. 5). Similarly, ASO 276-290 that disrupted ISTL2 but not ISTL1 had a smaller stimulatory effect on SMN2 exon 7 splicing (Fig. 5). It appears that the structural context set by TSL2, ISTL1, TSL3 and ISTL2 at the 5′ ss of exon 7 is designed to facilitate recruitment of negative regulators (such as hnRNP A1/A2B1) that somehow communicate with the factors interacting with C6U and A100G. This line of thoughts is consistent with the intron definition model that we recently proposed for the regulation of SMN exon 7 splicing.66 Unlike exon definition model that puts special emphasis on cis-elements within exon and flanking intronic sequences, intron definition model relies on regulatory sequences and structures within the entire intron. However, intron definition model is not complete till we fully understand how the 5′ ss of exon 7 communicates with the 3′ ss of exon 8. Similarly, we need to uncover the mechanism of by which pairing between the 5′ ss of exon 6 and the 3′ ss of exon 7 occurs. Reports that ASOs targeting Element 1 in intron 6 could stimulate SMN2 exon 7 underscores the role of intronic sequences upstream of exon 7.67 Further experiments are likely to uncover the role of additional intronic structures and transacting factors that will refine our understanding of the known interactions and reveal additional therapeutic targets.
Future perspectives
The biological role of SMN2, a gene specific to humans, remains elusive. Presence of SMN2 in humans is attributed to SMN gene duplication about 5 million years ago followed by a recent gene conversion that happened after human-chimpanzee divergence.68 No doubt, SMN2 serves as a promising target for SMA therapy by compounds that elevate the levels of SMN by enhancing SMN2 transcription and/or by correcting SMN2 exon 7 splicing. SMN2, which contributes towards the overall cellular pool of SMN, is likely to impact the severity of other neurodegenerative diseases. For instance, low SMN level in SMA has been found to downregulate α-synuclein, a protein associated with Parkinson’s disease.69 Several lines of evidence suggests that SMN modulates severity of Amyotrophic Lateral Sclerosis (ALS).70,71 We serendipitously discovered deletion of SMN2 in a patient cell line of Batten disease (BD).72 However, the significance of this finding is yet to be determined. The distinct splicing pattern of SMN2 under the conditions of oxidative stress makes SMN2 a stress sensing gene.72 Oxidative stress is a mediator of life history trade-offs.73 Therefore, SMN2 may have gained a role of a sensor that alerts and prepares the system for a better survival during stress-associated conditions.
Although skipping of SMN2 exon 7 is linked to the existing C6U and A100G mutations, many regulatory elements and transacting factors not associated with these positions have been reported.65 The antisense drug (ISIS-SMNRX based on ISS-N1 target) currently under phase 3 clinical trial, provides an undisputable proof that the desired splicing modulation could be achieved through intronic sequences.55 We and others have previously considered ISS-N1 as a portable linear cis-element, sequestration of which by an ASO produces the stimulatory effect (on SMN2 exon 7 splicing) by displacing a negative interacting factor. However, our recent finding of 10C-mediated LDI challenges this simplistic line of thoughts and supports the hypothesis that an ISS-N1-targeting ASO perturbs the inhibitory ISTL1 as well as makes the binding site of TIA1 accessible. Both, destabilization of ISTL1 and accessibility of TIA1 binding sites is conducive for the recruitment of U1 snRNP at the 5′ ss of exon 7. Similar outcome is expected to be produced by targeting ISS-N2 located in the middle of intron 7.
While our finding of the unique LDI that involves the deep intronic sequence is a major step forward towards understanding the mechanism of aberrant splicing in SMA, its implications go beyond SMA. A recent report connects an abrogation of an LDI within intron 3 of proteolipid protein 1 (PLP1) gene with X-linked leukodystrophy Pelizaeus-Merzbacher disease (PMD).74 This finding is just the tip of the iceberg as origins of many more genetic disorders associated with intronic mutations are yet to be established. LDIs could be loosely defined as high-order RNA structures formed with the help of secondary RNA structures. Although complementary sequences are the driving force of a LDI, a role of transacting factors in the formation and/or stabilization of a LDI could not be ruled out. In certain cases, a LDI could be enforced solely through protein-protein interactions.75 Now that we are beginning to appreciate the significance of intronic LDIs as the basis of diseases and their therapy, the stage is set for future studies to uncover the critical information present in secondary and high-order intronic structures that occupy a large portion of our transcriptome.
Acknowledgments
Authors acknowledge Joonbae Seo for critical reading of the manuscript. This work was supported by grants from National Institutes of Health (NIH) [NS055925, NS072259 and NS080294] and Salsbury Endowment (Iowa State University, Ames, IA, USA) (to RNS). ISS-N1 target (US patent # 7,838,657) was discovered in the Singh lab at UMASS Medical School (Worcester, MA, USA). Inventors, including RNS, NNS and UMASS Medical School, are currently benefiting from licensing of ISS-N1 target to ISIS Pharmaceuticals. Iowa State University holds intellectual property rights on GCRS and ISS-N2 targets. Therefore, inventors including RNS, NNS and Iowa State University could potentially benefit from any future commercial exploitation of GCRS and ISS-N2.
References
- 1.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McManus CJ, Graveley BR. RNA structure and the mechanism of alternative splicing. Curr Opin Genet Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mills JD, Janitz M. Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol Aging. 2012;33:1012.e1011. [Google Scholar]
- 5.Wan Y, Qu K, Ouyang ZQ, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. Genome-wide measurement of RNA folding energies. Mol Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat Rev Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 7.Palazzo AF, Gregory TR. The case for junk DNA. PLOS Genet. 2014;10:e1004351. doi: 10.1371/journal.pgen.1004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awano T, Kim JK, Monani UR. Spinal muscular atrophy: journeying from bench to bedside. Neurotherapeutics. 2014;11:786–795. doi: 10.1007/s13311-014-0293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivanesan S, Howell MD, Didonato CJ, Singh RN. Antisense oligonucleotide mediated therapy of spinal muscular atrophy. Transl Neurosci. 2013;4:1–7. doi: 10.2478/s13380-013-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo J, Howell MD, Singh NN, Singh RN. Spinal muscular atrophy: an update on therapeutic progress. Biochem Biophys Acta. 2013;1832:2180–2190. doi: 10.1016/j.bbadis.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell MD, Singh NN, Singh RN. Advances in therapeutic development for spinal muscular atrophy. Future Med Chem. 2014;6:1081–1099. doi: 10.4155/fmc.14.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton G, Gillingwater TH. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Nurputra DK, et al. Spinal muscular atrophy: from gene discovery to clinical trials. Ann Hum Genet. 2013;77:435–463. doi: 10.1111/ahg.12031. [DOI] [PubMed] [Google Scholar]
- 14.Bebee TW, Dominguez CE, Chandler DS. Mouse models of SMA: tools for disease characterization and therapeutic development. Hum Genet. 2012;131:1277–1293. doi: 10.1007/s00439-012-1171-5. [DOI] [PubMed] [Google Scholar]
- 15.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 16.Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, Brahe C, Saugier-Veber P, Bonnefont JP, Melki J. Refined characterization of the expression and stability of the SMN gene products. Am J Pathol. 2007;171:1269–1280. doi: 10.2353/ajpath.2007.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirth B, Brichta L, Hahnen E. Spinal muscular atrophy and therapeutic prospects. Prog Mol Subcell Biol. 2006;44:109–132. doi: 10.1007/978-3-540-34449-0_6. [DOI] [PubMed] [Google Scholar]
- 19.Kolb SJ, Battle DJ, Dreyfuss G. Molecular functions of the SMN complex. J Child Neurol. 2007;22:990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 20.Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiziano FD, Melki J, Simard LR. Solving the puzzle of spinal muscular atrophy: what are the missing pieces? Am J Med Genet Part A. 2013;161A:2836–2845. doi: 10.1002/ajmg.a.36251. [DOI] [PubMed] [Google Scholar]
- 22.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashima T, Rao N, Manley JL. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci USA. 2007;104:3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 27.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 29.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 30.Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, Manley JL, Sette C. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J. 2010;29:1235–1247. doi: 10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol Cell Biol. 2008;28:6929–6938. doi: 10.1128/MCB.01332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc Natl Acad Sci USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clery A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-β1. Nat Struct Mol Biol. 2011;18:443–450. doi: 10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- 34.Moursy A, Allain FH, Clery A. Characterization of the RNA recognition mode of hnRNP G extends its role in SMN2 splicing regulation. Nucleic Acids Res. 2014;42:6659–6672. doi: 10.1093/nar/gku244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho S, et al. hnRNP M facilitates exon 7 inclusion of SMN2 pre-mRNA in spinal muscular atrophy by targeting an enhancer on exon 7. Biochem Biophys Acta. 2014;1839:306–315. doi: 10.1016/j.bbagrm.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Cho S, et al. PSF contacts exon 7 of SMN2 pre-mRNA to promote exon 7 inclusion. Biochem Biophys Acta. 2014;1839:517–525. doi: 10.1016/j.bbagrm.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, Bridgeman SJ, Burghes AH, Kissel JT. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet. 2009;85:408–413. doi: 10.1016/j.ajhg.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vezain M, et al. A rare SMN2 variant in a previously unrecognized composite splicing regulatory element induces exon 7 inclusion and reduces the clinical severity of spinal muscular atrophy. Hum Mutat. 2010;31:E1110–1125. doi: 10.1002/humu.21173. [DOI] [PubMed] [Google Scholar]
- 39.Bernal S, et al. The c.859G>C variant in the SMN2 gene is associated with types II and III SMA and originates from a common ancestor. J Med Genet. 2010;47:640–642. doi: 10.1136/jmg.2010.079004. [DOI] [PubMed] [Google Scholar]
- 40.Miyajima H, Miyaso H, Okumura M, Kurisu J, Imaizumi K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J Biol Chem. 2002;277:23271–23277. doi: 10.1074/jbc.M200851200. [DOI] [PubMed] [Google Scholar]
- 41.Baughan TD, Dickson A, Osman EY, Larson CL. Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum Mol Genet. 2009;18:1600–1611. doi: 10.1093/hmg/ddp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irimura S, et al. HnRNP C1/C2 may regulate exon 7 splicing in the spinal muscular atrophy gene SMN1. Kobe J Med Sci. 2009;54:E227–236. [PubMed] [Google Scholar]
- 43.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh NN, Hollinger K, Bhattacharya D, Singh RN. An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing. RNA. 2010;16:1167–1181. doi: 10.1261/rna.2154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013;41:8144–8165. doi: 10.1093/nar/gkt609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyaso H, Okumura M, Kondo S, Higashide S, Miyajima H, Imaizumi K. An intronic splicing enhancer element in survival motor neuron (SMN) pre-mRNA. J Biol Chem. 2003;278:15825–15831. doi: 10.1074/jbc.M209271200. [DOI] [PubMed] [Google Scholar]
- 49.Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol. 2011;31:935–954. doi: 10.1128/MCB.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh NN, Androphy EJ, Singh RN. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem Biophys Res Commun. 2004;315:381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 51.Singh RN. Unfolding the mystery of alternative splicing through a unique method of in vivo selection. Front Biosci. 2007;12:3263–3272. doi: 10.2741/2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim SR, Hertel KJ. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J Biol Chem. 2001;276:45476–45483. doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 54.Keil JM, Seo J, Howell MD, Hsu WH, Singh RN, DiDonato CJ. A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy. Mol Ther Nucleic Acids. 2014;3:e174. doi: 10.1038/mtna.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castro D, Iannaccone ST. Spinal muscular atrophy: therapeutic strategies. Curr Treat Options Neurol. 2014;16:316. doi: 10.1007/s11940-014-0316-3. [DOI] [PubMed] [Google Scholar]
- 56.Young PJ, DiDonato CJ, Hu D, Kothary R, Androphy EJ, Lorson CL. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2β1. Hum Mol Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- 57.Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-β1. Hum Mol Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- 58.Bose JK, I, Wang F, Hung L, Tarn WY, Shen CK. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem. 2008;283:28852–28859. doi: 10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mende Y, Jakubik M, Riessland M, Schoenen F, Rossbach K, Kleinridders A, Köhler C, Buch T, Wirth B. Deficiency of the splicing factor Sfrs10 results in early embryonic lethality in mice and has no impact on full-length SMN/Smn splicing. Hum Mol Genet. 2010;19:2154–2167. doi: 10.1093/hmg/ddq094. [DOI] [PubMed] [Google Scholar]
- 60.Klar J, Sobol M, Melberg A, Mäbert K, Ameur A, Johansson AC, Feuk L, Entesarian M, Orlén H, Casar-Borota O, Dahl N. Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum Mutat. 2013;34:572–577. doi: 10.1002/humu.22282. [DOI] [PubMed] [Google Scholar]
- 61.Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5 splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 63.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kladwang W, VanLang CC, Cordero P, Das R. A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat Chem. 2011;3:954–962. doi: 10.1038/nchem.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wee CD, Havens MA, Jodelka FM, Hastings ML. Targeting SR Proteins Improves SMN Expression in Spinal Muscular Atrophy Cells. PLoS One. 2014;9:e115205. doi: 10.1371/journal.pone.0115205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh NN, Singh RN. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 2011;8:600–606. doi: 10.4161/rna.8.4.16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Osman EY, Miller MR, Robbins KL, Lombardi AM, Atkinson AK, Brehm AJ, Lorson CL. Morpholino antisense oligonucleotides targeting intronic repressor Element1 improve phenotype in SMA mouse models. Hum Mol Genet. 2014;23:4832–4845. doi: 10.1093/hmg/ddu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rochette CF, Gilbert N, Simard LR. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum Genet. 2001;108:255–66. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 69.Acsadi G, Li X, Murphy KJ, Swoboda KJ, Parker GC. Alpha-synuclein loss in spinal muscular atrophy. J Mol Neurosci. 2011;43:275–283. doi: 10.1007/s12031-010-9422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner BJ, Alfazema N, Sheean RK, Sleigh JN, Davies KE, Horne MK, Talbot K. Overexpression of survival motor neuron improves neuromuscular function and motor neuron survival in mutant SOD1 mice. Neurobiol Aging. 2014;35:906–915. doi: 10.1016/j.neurobiolaging.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamazaki T, Chen S, Yu Y, Yan B, Haertlein TC, Carrasco MA, Tapia JC, Zhai B, Das R, Lalancette-Hebert M, Sharma A, Chandran S, Sullivan G, Nishimura AL, Shaw CE, Gygi SP, Shneider NA, Maniatis TT, Reed R. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2:799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh NN, Seo J, Rahn SJ, Singh RN. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS One. 2012;7:e49595. doi: 10.1371/journal.pone.0049595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monaghan P, Metcalfe NB, Torres R. Oxidative stress as a mediator of life history trade-offs: mechanisms, measurements and interpretation. Ecol Lett. 2009;12:75–92. doi: 10.1111/j.1461-0248.2008.01258.x. [DOI] [PubMed] [Google Scholar]
- 74.Taube JR, Sperle K, Banser L, Seeman P, Cavan BC, Garbern JY, Hobson GM. PMD patient mutations reveal a long-distance intronic interaction that regulates PLP1/DM20 alternative splicing. Hum Mol Genet. 2014;23:5464–5478. doi: 10.1093/hmg/ddu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:172–185. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]