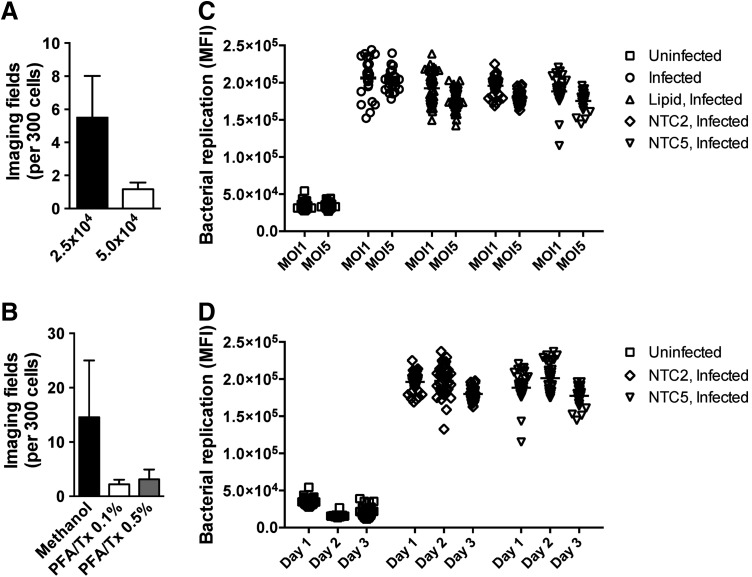
Fig. 3.
Identification of optimal experimental parameters for image-based quantification of bacterial replication. (A) The average number of imaging fields required to identify at least 300 THP1 cells plated at different cell densities per well of a 96-well plate and infected with B. cenocepacia for 24 h at MOI 1. (B) The average number of imaging fields required to identify at least 300 THP1 cells plated at 5 × 104 cells per well of a 96-well plate, infected with B. cenocepacia for 24 h at MOI 1, and subjected to different fixation and permeabilization conditions. (C) THP1 cells were infected with B. cenocepacia at MOI 1 or MOI 5 for 24 h, and bacterial replication was quantified by imaging as described in the Materials and Methods section. Before infection, cells were treated with either HiPerFect transfection lipid or transfected with nontargeting control (NTC) siRNA where indicated. (D) THP1 cells were infected with B. cenocepacia at MOI 1 for 24 h, and bacterial replication was quantified by imaging as described in the Materials and Methods section. Data are shown from infections conducted on 3 separate days. Before infection, cells were transfected with NTC siRNA where indicated. Data shown are representative of two independent experiments for (A) and (B) (mean + SD) and (C) Individual data points from 24 to 48 wells of a 96-well plate. (D) Individual data points from 32 separate wells for each condition from three replicate experiments.