Abstract
Mesenchymal stem cells promising role in cell-based therapies and tissue engineering appears to be limited due to a decline of their regenerative potential with increasing donor age. Six age groups from bone marrow mesenchymal stem cells of Wistar rats were studied (newborn, infant, young, pre-pubertal, pubertal and adult). Quantitative proteomic assay was performance by iTRAQ using an 8-plex iTRAQ labeling and the proteins differentially expressed were grouped in pluripotency, proliferative and metabolism processes. Proliferation makers, CD117 and Ki67 were measure by flow cytometry assay. Real time polymerase chain reaction analysis of pluripotency markers Rex1, Oct4, Sox2 and Nanog were done. Biological differentiation was realized using specific mediums for 14 days to induce osteogenesis, adipogenesis or chondrogenesis and immunostain analysis of differentiated cell resulting were done. Enzimoimmunoassay analysis of several enzymes as L-lactate dehydrogenase and glucose-6-phosphate isomerase were also done to validate iTRAQ data. Taking together these results indicate for the first time that mesenchymal stem cells have significant differences in their proliferative, pluripotency and metabolism profiles and those differences are age depending.
Mesenchymal Stem Cells (MSCs) have self-renewal capacity and multiple differentiation potentials, and a priori, could play important roles in regenerative medicine. The promising role of MSCs in cell-based therapies is their trophic, paracrine and immunomodulatory functions that may have the greatest therapeutic impact in vivo1,2. Tissue engineering from MSCs are of highly importance for regeneration of mesenchymal tissues such as craniofacial bone3, cartilage4 and connective tissues5.
Since several years ago, numerous studies have shown that MSCs from different tissues have similar levels of surface antigen expression, immunosuppressive activity, and differentiation ability6. The ability of MSC to carry out normal tissue regeneration in the body and their potential for using in clinical applications may be impaired by loss of stem cell number and function with age7. There are different rate of cell proliferation and clonality between MSCs depending of source from the cells are obtained8 and the chronological age form the donors and also the number of the in vitro culture passages9. MSCs are missing their characteristics during the chronological or in vitro culture of them10 but not in the same way. Various attempts have been made to address challenges associated with aging of MSC including culture in hypoxic conditions11 and ectopic expression of pluripotency-associated factors12. This study applied the 8-plex iTRAQ system to analyze MSCs from six different aging groups, as this quantitative proteomic technology has the capability to compare several time points in a single experiment.
The major contributor to the development of the senescent cellular phenotype is hyper activation of nutrient sensor and growth pathways, in particular mTOR and its derivative complexes mTORC1 and mTORC213,14. mTOR family regulates senescence and autophagy during reprogramming of somatic cells to pluripotency indicating the important role of energy metabolism to stem cell renewal and aging15. We studied the relationship between mTOR and the proliferation markers CD117 and Ki67 using imatinib mesylate, the inhibitor of tyrosine kinase receptor for CD11716 and JK184 which reduce expression of Ki6717. We establish a framework for future comparative and functional studies; we have analyzed the phenotypic, genotypic features and biological-related changes in MSCs of rat bone marrow from animals of different ages.
Material and Methods
Isolation and culture of cells
For isolation of MSCs, the animals were anesthetized with Fluorane (Izasa, A Coruña, SP) and sacrificed by cervical dislocation method. Femurs were dissected from male Wistar rat (Animal Service, CHUAC) at different ages: neonate (0 days old), infant (7 days old), young (14 days old), pre-pubertal (35–38 days old), pubertal (45 days old) and adult (2 months old). All the methods were carried out in “accordance” with the approved guidelines of Spanish law (32/2007). All experimental protocols were approved by Animal Ethical Committee of Galicia. The protocol used by Karaoz et al.18 was followed in this work. Briefly, the ends of the bones were cut away and a 21-gauge needle that was inserted into shaft of the bone marrow was extruded by flushing with 5 ml D-Hank’s solution supplemented with 100 IU/ml penicillin–100 mg/ml streptomycin (all from Life Technologies, Madrid, Spain). Marrow plug suspension was dispersed by pipetting, successively filtered through 70-μm mesh nylon filter (BD Biosciences, Bedford, MA, USA), and centrifuged at 20000 g for 10 min. Supernatant containing thrombocytes and erythrocytes was discarded, and the cell pellet was resuspended in the medium. The cells from four rats were seeded onto 100 cm2 dish plate (TM Nunclon) and incubated at 37 °C with 5% humidified CO2. The MSCs were isolated on the basis of their ability to adhere to the culture plates. On the third day, red blood cells and other non-adherent cells were removed and fresh medium was added to allow further growth. The adherent cells grown to 70% confluence were defined as passage zero (P0) cells. After 5 min of centrifugation, 1 × 106 MSCs were seeded on two dish plates 100 cm2 (TM Nunclon) in RPMI supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin (all from Sigma-Aldrich, St. Louis, MO, USA). The medium was added and replaced every 3 or 4 days for 2 weeks. Before being used in inducing mesoderm differentiation, the MSCs had been expanded for 2 passages and characterized. Afterwards MSCs from adult group were incubated during 12 hours previously to protein extraction with imatinib mesylate or JK184 (all from Sigma-Aldrich).
Flow Cytometry
To characterize the different populations of MSCs from chronological different animals, their MSCs were washes twice in PBS (Sigma-Aldrich, St. Louis, MO, USA), then pre-blocked with 2% rat serum in PBS. The following direct antibodies were used: PE-conjugated mouse anti-human CD34 (1:20 from DakoCytomation, Barcelona, SP); FITC-conjugated mouse anti-rat CD45 (1:20 BD Pharmingen, New Jersey, USA); PE-Cy5.5-conjugated mouse anti-rat CD90 (1:20 Immunostep, Salamanca, SP) and APC-conjugated mouse anti-rat CD29 (1:20 Immunostep, Salamanca, SP). The cells were washed with PBS after one hour of incubation with the corresponding antibody at room temperature. To check proliferation profile of the different populations of MSCs from chronological different animals, their MSCs were incubated with APC conjugated mouse anti-rat CD117 (1:20 Immunostep, Salamanca, SP), mouse anti-BRDU (sigma-Aldrich) and FITC conjugated mouse anti-human Ki67 (1:20 Immunostep, Salamanca, SP). The secondary FITC-conjugated rabbit anti-mouse antibody was used to link cells incubated with anti-BRDU. The stained cells were then washed twice with PBS and 2 × 105 cells were analyzed with a FACSAria flow cytometer (BD Science, Madrid, SP). FACS data was generated by DIVA software (BD Science). Negative control staining was performed using FITC-conjugated mouse IgG1K isotype, PE- conjugated mouse IgG1K isotype, PE-Cy5.5- conjugated mouse IgG1K isotype and APC- conjugated mouse IgG1K isotype (all from BD Pharmingen). For the intracellular ROS accumulation was used H2DCFDA. Upon oxidation by ROS, the non-fluorescent H2DCFDA is converted to the highly fluorescent 2′,7′-dichlorofluorescein. MitoSOX™ Red Reagent was used to determine mitochondrial ROS including superoxide dismutase activity. Tetramethylrhodamine, methyl ester (TMRM) (from Thermo Fisher Scientific, Life Technologies, SP), the permanent dye that accumulates in active mitochondria with intact potentials, was used to detect functional mitochondria in the MSCs at different ages following functional mitochondrial staining protocol from commercial.
Proliferation assay
Different numbers of cells (0, 1000, 2000, 4000, 8000 and 16000 cells), were plated in triplicate in 96-well plates and allowed to adhere for 8 hours, were used to calculate the proliferation curve. The number of cells was then calculated using CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI, USA) following manufacturer instructions. For the assay, 4000 cells were plated for each cell line in triplicate in 96-well plates, and the total number of cells was calculated at different time points (0, 1, 2, 5 and 6 days).
The cytotoxicity assay was realized using Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc. MD 20850, USA). Briefly 100 all of cell suspension (5000 cells/well) in a 96-well plate were incubated for 24 hours at at 37 °C, 5% CO2. After that medium with imatinib mesylate or JK184 was added to the wells 48 hours later, 10 μl of CCK-8 solution was added to each well of the plate. After four hours incubation the absorbance was measured at 450 nm using a microplate reader.
Histological and immunohistochemical analysis
MSCs following differentiation into chondrocyte-like or adipocyte-like cells were frozen in OCT embedding matrix (BDH Chemicals, Poole, UK). The cells were fixed in 4% (w/v) paraformaldehyde (Sigma-Aldrich) in PBS at pH 7.6. Some cells were stained with Safranin O, Modified Masson´s (Sigma-Aldrich) to evaluate the distribution of proteoglycan in the extracellular matrix generated by the cells differentiated towards chondrocyte-like cells. Other cells were stained with Alizarin red (Sigma-Aldrich) to check alkaline deposits in cells differentiated towards osteocyte-like cells. Other cells were stained with Oil red (Sigma-Aldrich) to check oil drops in cells differentiated towards adipocyte-like cells.
Densitometry analysis
AnalySIS Image Processing (Soft Imaging system GmbH V. 5.0, Olympus, Münster, Germany) was used to do a densitometry quantification of the staining obtained by immunohistochemistry analysis shown in the plots. Three fields 200 mm2 in size from each inmunostain- safranine O, oil red, modified Masson’s and alizarin red - and time studied were quantified using arbitrary units for immunohistochemistry values provided by the computer program. Values expressed as percentage of positive stain for each marker studied were used for immunohistochemistry analysis. All values were referenced with respect to values obtained from cells cultured in the control medium (RPMI 5% knockout serum, 1% penicillin and 1% streptomycin).
Real time quantitative polymerase chain reaction (qRT-PCR) analysis
Primers for amplification of rat Rex1, Oct4, Sox2 and Nanog genes were used to determine the expression of those markers for pluripotency in the different populations of MSCs from chronological different animals. Details are shown in Table 1. The amplification program consisted of initial denaturation at 92 °C for 2 minutes followed by 40 cycles from 92 °C for 15 seconds, annealing at 55–62 °C, depending on the gene, for 30 seconds and extension at 72 °C for 15 seconds. PCR analyses were done in triplicate, with each set of assays repeated three times. To minimize the effects of unequal quantities of starting RNA and to eliminate potential sources of inconsistency, relative expression levels of each gene was normalized to ribosomal protein (HPRT) using the 2-ΔΔCt method19. Control experiments utilized no reverse transcriptase.
Table 1. Specific primers for real-time reverse transcriptase-polymerase chain reaction (RT-PCR) amplification, listed with their annealing temperature (A.T.).
| Gene Name | Fw primer | Rv primer | mRNA ID | A.T. (°C) |
|---|---|---|---|---|
| Rex1 | gtgcatcacacctcagactgt | cgttggttgaaggccaactg | NM_005106.4 | 61 |
| Oct4 | ctcctggagggccaggaatc | atatacacaggccgatgtgg | NM_00510 | 61 |
| Sox2 | ctccgggacatgatcagc | ggtagtgctgggacatgtgaa | NM_001109181.1 | 61 |
| Nanog | atgcctcacacggagactgt | aagtgggttgtttgcctttg | NM_005103.4 | 61 |
| Vinculin | aggagaccttgcgaagacagg | gcggttgccacttgtttag | NM_001107248 | 61 |
| HPRT | agccgaccggttctgtcat | agccgaccggttctgtca | NM_012583.2 | 61 |
Fw = forward; Rv = reverse.
Protein isolation and immunoblot analysis
Immunoblot analysis was performed on 40 μg of total protein extracted from MSCs, as previously described20. The blots were probed with antibodies, made into rabbit, directed against mTOR, raptor (Cell Signaling, Izasa, Madrid, ES), vimentin, superoxide dismutase (SOD-2), lamin A/C and tubulin (all form Sigma-Aldrich) was used for housekeeping. A secondary anti-goat antibody (Cell Signaling) was used to visualize proteins using an AmershamTM ECLTM Western Blotting Analysis System (GE Healthcare, Amersham Biotechnology, Manchester, UK). Ideal concentrations for each antibody were determined empirically. Working concentrations were 1:1000 of the recommended stock solutions.
iTRAQ labelling
Equal amounts of proteins from each group of different age cells (100 μg) were denatured with 2% sodium dodecyl sulfate (SDS) in 1 M tryethylammonium bicarbonate (TEAB) (ABSciex, Foster City, CA). The samples were then reduced for 1 h at 60 °C using 50 mM tris-(2-carboxyethy) phosphine (TCEP) (ABSciex), and cysteine-blocked with 84 mM iodoacetamide (Sigma-Aldrich) at room temperature in the dark for 30 min. The proteins were digested with spectrometry grade trypsin (Gold Mass, Promega, Madison,WI) at a concentration of 1:50 trypsin/protein for 16 h at 37 °C. Each peptide solution was labeled for 1.5 h at room temperature using the iTRAQ reagents previously reconstituted in 70 μL of ethanol, following the manufacture Protocol (ABSciex). The samples were labeled with iTRAQ reagents as follows: newborn: 119 and 121 as a control infant: 114; young: 116; pre-pubertal: 118; pubertal: 115; adult: 117. The reaction was stopped by adding deionized water, and the labeled samples were combined. The mixture was desalted using home-made stage-tips.
iTRAQ relative quantification by 2D-LC-MALDI-TOF/TOF analysis
In a first step, the desalted peptides were fractionated by basic reversed phase extraction in a 1400 HPLC system (Agilent). The fractions were collected along a 110 minutes gradient and subjected to further acidic reversed phase extraction in a nanoHPLC system (Tempo, ABSciex) into a C18 silica-based column (New Objective, Woburn, MA) with an internal diameter of 300 Ả. The injection volume was 5 μL, and peptides were eluted during a ninety minutes gradient at a constant flow rate of 0.35 μL/min. Eluting peptides were automatically mixed with alpha-cyano at 4 mg/mL en 70% AcN, TFA 0.1% and deposited on a MALDI LC-plate using a SunCollect (SunChrom) spotter. The chromatograms, composed by 350 spots, each one comprising a 15 sec deposition, were then analyzed in a 4800 MALDI-TOF/TOF platform (ABSciex). 4000 series Explorer v.4.2 software was used to generate the spectra and peak list. After manual deposition of mass calibrates, plate model and default calibration of the MALDI plate was done with a laser voltage of 3200 kV and 1000 shots/spectrum. Samples were automatically analyzed in MS mode with a laser voltage of 3400 kV and 1500 shots/spectrum. Automated precursor selection was done using a Job-wide interpretation method (up to 12 precursors/fraction, Signal to Noise lower threshold = 50) excluding trypsin autolytic peptides and other background ions, with a laser voltage of 4200 and 2000 shots/spectrum. CID collision energy range: medium. LC-MALDI-TOF/TOF data were analyzed using ProteinPilot 4.0 software (ABSciex). Protein Pilot Search parameters were as follows: Sample type: iTRAQ 8-plex; Cys-alkylation: iodoacetamide; Digestion: trypsin; ID focus: Biological modifications; Database: last SwissProt release; Species filtering: none; Search effort: Thorough ID and Detection Protein Threshold Unused ProtScore (Conf)>1.3 (95.0%). Scoring model was defined by the Paragon algorithm. In the case of the high complexity samples, False Discovery Rate -FDR- was estimated in less than 1% by doing the searching in parallel against a decoy database using “PSPEP on” mode -data not shown-.
Bioinformatics
Biological functional analysis of different modulated proteins detected by iTRAQ quantification, were categorized according to their function, biological process and cellular component, using the String 9.0 software21. Proteins with statistically significant changes were identified by filtering according to these criteria: 1) they had to be present in two biological replicates; 2) changes between groups had to be statistically significant (P < 0.05); and 3) fold change had to be greater than 1.2 and lower than 0.8 (date do not shown) This approach allowed us to select 201 differentially expressed proteins for further analysis.
Enzymatic Analysis
5 × 105 cells from each group of different age were used for the assessment of enzyme activities. The cells were homogenized in 200 μL of 250 mM sucrose(Sigma-Aldrich, St.Louis, MO), 50 mM HEPES (Sigma-Aldrich), 0,5 mM EDTA (Sigma Aldrich) and one tablet protease inhibitor cocktail (Roche, Mannheim, Germany ). Enzymes activities were determined using a SUNRISE spectrophotometer (TECAN, Mannedorf, Switzerland). Reaction rates of enzymes were determined by the increase or decrease in absorbance of NAD(P)H (Sigma-Aldrich, St.Louis, MO) at 340 nm at 37 °C. Lactate dehydrogenise (EC 1.1.1.27) was determined in BM-MSCs using 50 mM Trizma base (pH 7,4), 0,15 mM NADH and 5 mM sodium pyruvate (omitted for control) (all Sigma Aldrich, St.Louis, MO). Glucose-6-phosphate 1-deshydrogenase (EC 1.1.1.49) and 6-phosphogluconate dehydrogenase, decarboxylating (EC 1.1.1.343) was determined in BM-MSCs using 78 mM Trizma base, 5 mM MgCl2(pH 7,4), 0,1 mM NADP, 0,5 mM D-Glucose 6-phosphate disodium salt hydrate and 6-Phosphogluconic acid trisodium salt (omitted for control) (all Sigma-Aldrich, St.Louis, MO).
Statistics
All experiments were performed in triplicate and one representative is shown. Non-parametric statistical analyses were performed by Mann-Whitney-U and Kruskal-Wallis tests using GraphPad Prism6 (GraphPad Software, La Jolla, CA). Each group was compared with previous group. A p value less than 0.05 or 0.01 were considered statistically significant. All the the data are presented as standard error of the mean.
Results
Characterization of populations of MSCs from different ageing group by flow cytometry reveled that no statistical significant differences exist between group respects levels of mesenchymal and hematopoietic markers used for that (Fig. 1A). Positive cells for CD45 and CD34 were less than 1%, positive cells for CD29 were 30 ± 5% and positive cells for CD90 were 75 ± 5% in all groups studied.
Figure 1. Proliferation profile from rat mesenchymal stem cells at different age.
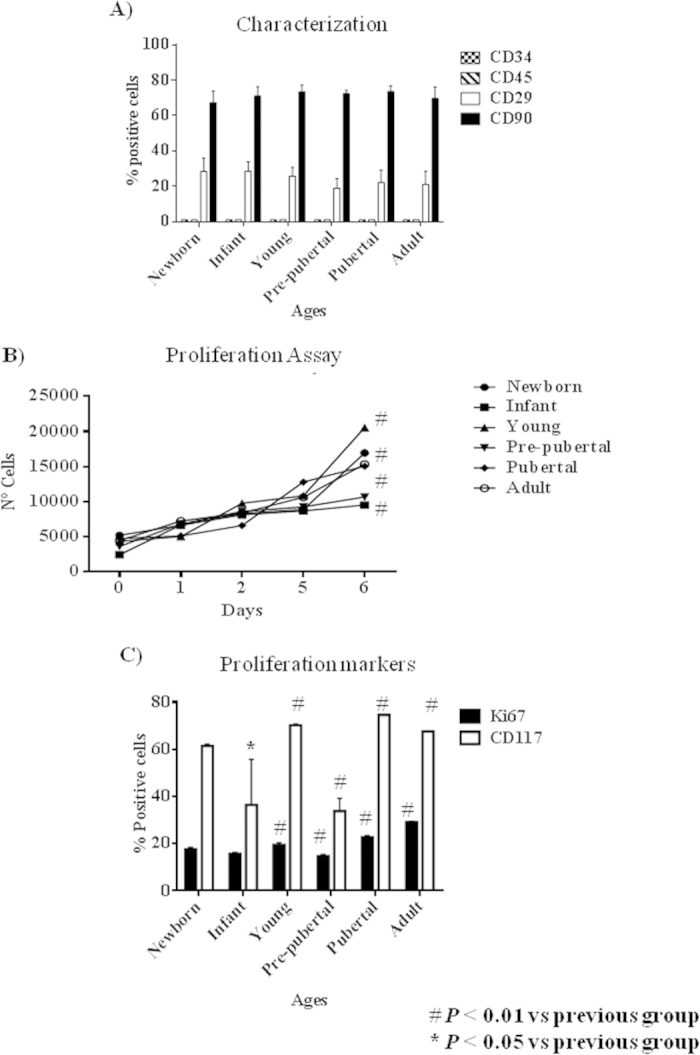
(A) Characterization by flow cytometry assay of percentage of positives mesenchymal stem cells markers (CD29 and CD73) and negative hematopoietic markers (CD34 and CD45). (B) Proliferation assay of studied aging groups for 6 days. (C) Percentage of proliferation markers, DC117 and Ki67, from studied aging groups by flow cytometry assay. One representative experiment is shown. #p value less than 0.05 compared with previous group and *p value less than 0.01 compared with previous group, were considered statistically significant using Mann-Whitney-U tests.
Proliferation Assays results indicated that MSCs from groups of newborn (17 × 103 ± 100), young (21 × 103 ± 200), pubertal (15 × 103 ± 300) and adult (16 × 103 ± 100) animals had a statistically significant higher (p < 0.01) number the cells compared to infant (9 × 103 ± 500), and pre-pubertal (10 × 103 ± 500) (Fig. 1B).
Flow cytometry assays to detect CD117 and Ki67 positive cells indicated that MSCs from pubertal and young groups had the statistical significant (p < 0.01 and p < 0.05 respectively)) higher CD117 positive cells percentage of MSCs (74.65 ± 0.07 and 71.95 ± 3.10 respectively) than the rest of the groups studied, newborn: 61.53 ± 0.37; infant: 60.50 ± 1.58; adult: 61.12 ± 6.35 and pre-pubertal: 35.25 ± 2.14. On the other hand, infant and pre-pubertal groups had the statistical significant (p < 0.05) lower Ki67 positive cells percentage (15.63 ± 0.24 and 14.65 ± 0.41 respectively) than the rest of the groups studied, newborn: 18 ± 0.55; young: 19.33 ± 0.43; pubertal 22.68 ± 0.40 and adult: 29.02 ± 0.16 (Fig. 1C)
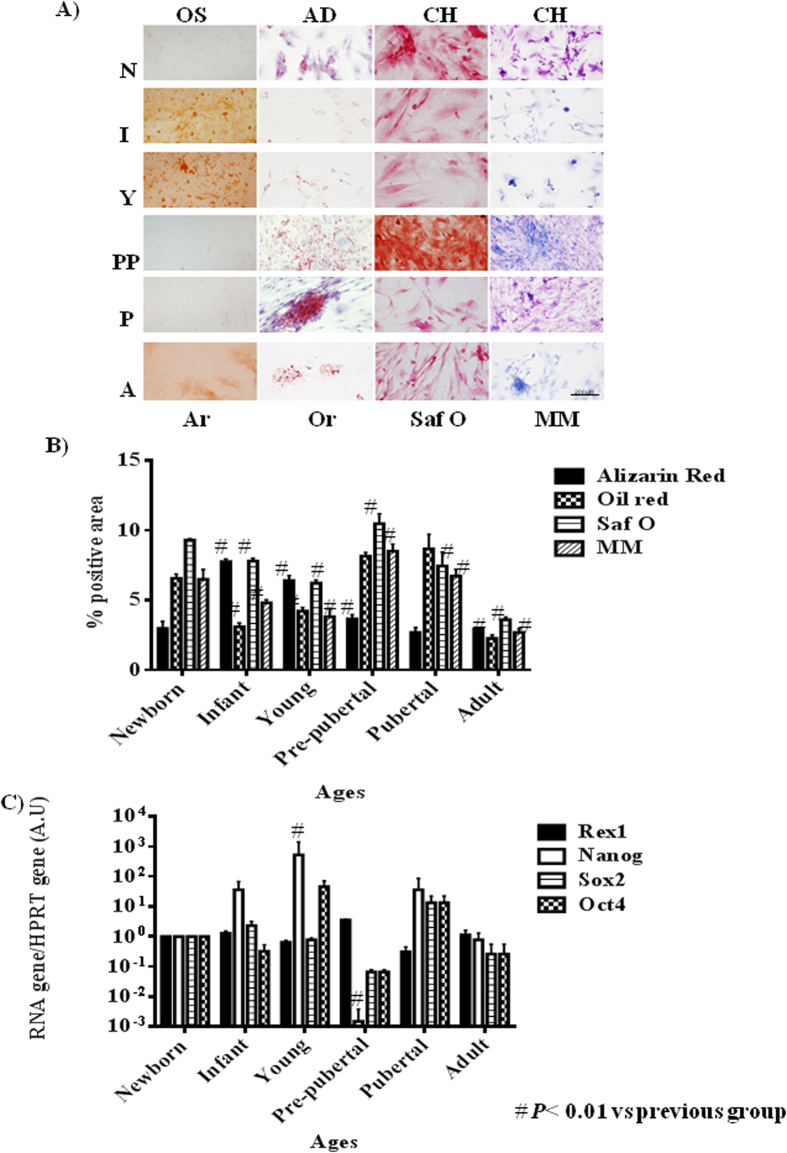
Differentiation capacity of the groups studied was tested through direct mesoderm induction using specific culture medium. It was observed that pre-pubertal group presented statistically significant (p < 0.05) highest stain for safranine O, modified Masson´s and oil red by histological analysis followed by pubertal with respect to the other groups. Young group presented the highest staining, statistically significant (p < 0.05) for alizarin red with respect to others groups and the adult group presented the lowest statistically significant (p < 0.05) differentiation potential with respect to other groups (Fig. 2A,B). Nanog, Oct4, Sox2 and Rex1 gene expression were tested by qRT-PCR analysis to check the pluripotency potential of the studied groups. The results shown the statistically significant (p < 0.05) highest expression of Nanog in young group with respect to the others groups in opposition of statistically significant (p < 0.01) decrease expression of this same gene, Nanog, in the pre-pubertal group respect to the others (Fig. 2C).
Figure 2. Pluripotency profile from rats mesenchymal stem cells at different age.
(A) Representative pictures of mesenchymal stem cell from bone marrow of studied age group after 14 days with specific differentiation medium. On the top specific medium is indicated; OS=osteogenic medium; AD = adipogenic medium; CH = chondrogenic medium. Differentiation medium are indicated in Material and methods. On the bottom stain is indicated; Ar = alizarin red; Or = oil red; Saf O = safranine O and MM = Modified Masson’s stain. Straight size is 200 μM. (B) Densitometry study of mesenchymal stem cell from bone marrow of studied aging group after 14 days with specific differentiation medium after immunostaining assay. AnalySIS Image Processing computer was used to quantify the signal of different stain obtained. #p value less than 0.01 was considered statistically significant using Mann-Whitney-U tests. (C) Histogram represents gene expression of pluripotency markers, Rex1, Nanog, Sox2 and Oct4. Real-time reverse transcriptase PCR (qRT-PCR) analysis normalized by expression of HPRT gene used as housekeeping. #p value less than 0.01 compared with previous group was considered statistically significant using Mann-Whitney-U tests. Three replicates were made.
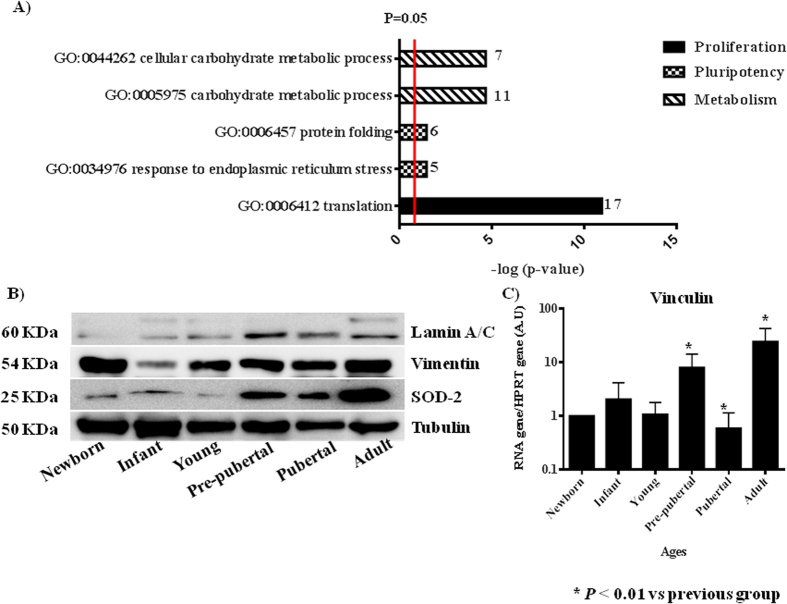
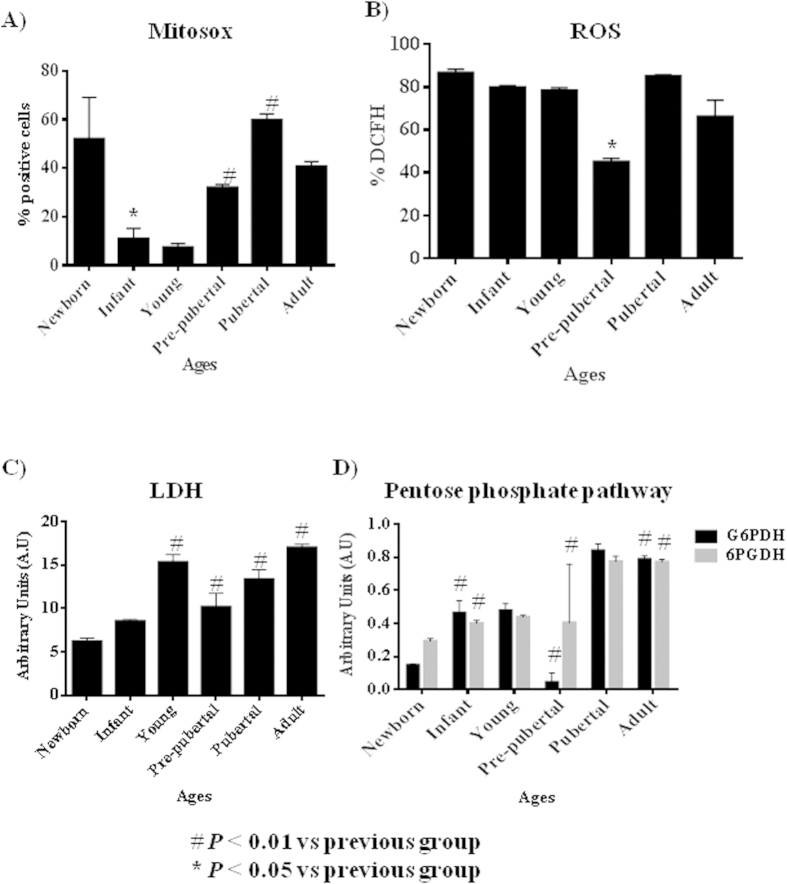
All proteins from MSCs of rat bone marrow at different ages studied were compared between them. Summary each group was composed of a pool from 6 animals and two different iTRAQ experiments were performed. The results obtained in the iTRAQ study indicated that 1.072 proteins were identified, 201 of them statistically significant modulated between groups (Table 2). These proteins have been grouped by three processes attending String 9.0 software; those groups were proliferation (60 proteins), pluripotency (86 proteins) and energy metabolism (55 proteins) (Table 2). Significant activates pathways obtained by comparing modulated proteins obtained by iTRAQ analysis employing functional annotations according to the String 9.0 software and classified in three biological process for better comprehension were shown in Fig. 3. A. Several proteins found in our analysis associated with proliferation were 60Sribosomal proteins with different sedimentation speed like 60S RP L10, 60S RP L9, 60S RP L23, 60S RP L24, 60S RP L4, 60S RP L6 and 60S RP L7; also Vinculin which gene expression was validated by qRT-PCR analysis (Fig. 3D), all of them were statistically significant (p < 0.05) higher in newborn and adults with respect to the others groups. Superoxide dismutase-2 (SOD2) and Lamin A were increasing through the increasing age group like occurred in the iTRAQ analysis; all of them were validated by western blot (Fig. 3B). Mitosox and total ROS were studied by flow cytometry to explain SOD2 results in our iTRAQ study, ROS and mitosox were statistically significant (p < 0.05) lower in infant and pre-pubertal groups with respect to the others groups respectively (Fig. 4A,B). A fluorescence-based assay to detect functional mitochondria indicated that adult group had their functional mitochondria statistically significant decreased with respect to others groups of study (Fig. 5B). Proteins found in our iTRAQ analysis were associated with pluripotency like histones H1.5; H2B; H4 and protein disulfide-isomerase A1 (PDIA1) which were statistically significant (p < 0.05) high-regulated in infant and pubertal groups with respect to the others (Table 2). Several proteins found in our iTRAQ analysis associated whit energy metabolism were lactate dehydrogenase (LDH), glucose 6 phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH), which were validated through analysis of their activity by enzimoimmunoassay (GPI). Lactate dehydrogenase (LDH) activity was increased in young group in front to infant and newborn group, decreased in pre-pubertal group in a statistically significant way (p < 0.01) and come back to increase its activity in pubertal and adult groups (Fig. 4C), glucose 6 phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH) were statistically significant increased (p < 0.01) in pubertal and adult groups in front to others (Fig. 4D).
Table 2. List of modulated proteins in mesenchymal stem cells at different ages classified according to their principal biological process using iTRAQ analysis.
| Accession | Name | Peptides(95%) | I/N | PVal I/N | Y/I | PVal Y/I | PP/Y | Pval PP/Y | P/PP | PVal P/PP | A/P | PVal A/P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolism | ||||||||||||
| Q6P783 | 6-phosphofructokinase | 5 | 0,7228 | 0,1496 | 1,0497 | 0,7085 | 1,1027 | 0,3079 | 1,4117 | 0,0388 | 1,2082 | 0,2192 |
| Q7TP11 | 6-phosphogluconate dehydrogenase, decarboxylating | 5 | 0,8664 | 0,0404 | 0,9404 | 0,345 | 1,1936 | 0,1035 | 1,2877 | 0,0026 | 0,6857 | 0,0099 |
| P06761 | 78 kDa glucose-regulated protein | 35 | 0,9172 | 0,0089 | 0,9677 | 0,334 | 1,1997 | 0 | 0,8295 | 0,002 | 1,4611 | 0 |
| M0RDC5 | Acyl-CoA-binding protein (Fragment) | 1 | 0,9923 | 0,9393 | 1,0172 | 0,8471 | 1,0129 | 0,8865 | 1,1744 | 0,4947 | 1,408 | 0,0299 |
| F1LN88 | Aldehyde dehydrogenase, mitochondrial | 9 | 0,915 | 0,2981 | 0,9521 | 0,605 | 1,0255 | 0,8092 | 0,8105 | 0,1287 | 1,249 | 0,0303 |
| P07943 | Aldose reductase | 9 | 1,0984 | 0,3717 | 0,8893 | 0,186 | 0,9549 | 0,5728 | 0,9715 | 0,7164 | 1,2055 | 0,0389 |
| Q91W30 | Aldose reductase-like protein | 10 | 1,6948 | 0,0054 | 0,5021 | 0,0183 | 1,2079 | 0,2492 | 1,0106 | 0,9424 | 1,2476 | 0,0229 |
| D3ZUM4 | Beta-galactosidase | 5 | 1,0315 | 0,7835 | 1,3171 | 0,0441 | 1,0942 | 0,385 | 1,1471 | 0,1506 | 0,9005 | 0,1892 |
| O35567 | Bifunctional purine biosynthesis protein PURH | 13 | 0,9914 | 0,9389 | 0,8798 | 0,2663 | 1,1514 | 0,3918 | 0,8194 | 0,0166 | 1,1312 | 0,0362 |
| Q99JD5 | Branched-chain-amino-acid aminotransferase | 5 | 1,0463 | 0,7187 | 0,842 | 0,0756 | 1,3485 | 0,0127 | 0,9146 | 0,4169 | 0,9297 | 0,5047 |
| P15791 | Calcium/calmodulin-dependent protein kinase type II subunit delta | 5 | 0,9646 | 0,825 | 0,9676 | 0,8106 | 1,3814 | 0,0071 | 0,7966 | 0,044 | 1,0233 | 0,8049 |
| G3V9E3 | Caldesmon 1, isoform CRA_b | 18 | 1,0648 | 0,1599 | 0,9241 | 0,1919 | 1,4728 | 0 | 0,7388 | 0,0002 | 1,6353 | 0 |
| Q08290 | Calponin-1 | 9 | 0,8714 | 0,0531 | 1,3003 | 0,0464 | 0,8214 | 0,2876 | 0,723 | 0,0013 | 1,2572 | 0,0412 |
| P37397 | Calponin-3 | 10 | 1,3337 | 0,0378 | 0,8281 | 0,0224 | 1,0857 | 0,2275 | 0,7288 | 0,0035 | 1,1632 | 0,1397 |
| P18418 | Calreticulin | 12 | 1,1514 | 0,0446 | 0,8607 | 0,0394 | 1,3403 | 0,0004 | 0,7119 | 0,0002 | 1,4109 | 0,0004 |
| G3V6S3 | Calumenin | 5 | 1,0591 | 0,4369 | 1,0768 | 0,3271 | 0,9833 | 0,8456 | 0,7673 | 0,2524 | 1,5598 | 0,0141 |
| Q6P6T6 | Cathepsin D | 7 | 0,8801 | 0,1287 | 1,0622 | 0,3991 | 1,5419 | 0,0105 | 0,9111 | 0,1808 | 1,3326 | 0,0234 |
| P97601 | Chaperonin 10 | 3 | 1,0204 | 0,7796 | 1,0011 | 0,9902 | 0,9415 | 0,5177 | 0,939 | 0,4419 | 1,4235 | 0,0087 |
| G3V936 | Citrate synthase | 3 | 0,7934 | 0,0267 | 1,167 | 0,0848 | 0,7904 | 0,0267 | 1,1452 | 0,2405 | 0,857 | 0,2318 |
| F1M779 | Clathrin heavy chain | 9 | 1,2157 | 0,0137 | 1 | 0,9994 | 0,9612 | 0,7295 | 1,1343 | 0,2338 | 0,8853 | 0,3235 |
| Q6TUH9 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | 3 | 0,7341 | 0,2686 | 1,3756 | 0,2277 | 1,6421 | 0,0328 | 1,9857 | 0,0672 | 0,7338 | 0,0642 |
| P47875 | Cysteine and glycine-rich protein 1 | 7 | 1,0015 | 0,9815 | 1,0448 | 0,5131 | 1,2576 | 0,0488 | 0,6896 | 0,0389 | 1,2501 | 0,0705 |
| O08651 | D-3-phosphoglycerate dehydrogenase | 3 | 0,8786 | 0,1902 | 0,7465 | 0,0308 | 1,1981 | 0,1105 | 1,1565 | 0,1448 | 1,0565 | 0,5069 |
| Q5BJ93 | Enolase 1, (Alpha) | 23 | 1,0226 | 0,655 | 1,026 | 0,5722 | 1,054 | 0,2406 | 0,7412 | 0 | 1,2752 | 0,0123 |
| Q8R4A1 | ERO1-like protein alpha | 5 | 0,8236 | 0,0936 | 1,0042 | 0,9607 | 1,1541 | 0,1632 | 1,0604 | 0,6296 | 1,4685 | 0,0439 |
| P05065 | Fructose-bisphosphate aldolase A | 7 | 0,7852 | 0,0001 | 0,9961 | 0,9275 | 1,1519 | 0,0055 | 1,17 | 0,0228 | 1,1494 | 0,0589 |
| P11762 | Galectin-1 | 13 | 0,8947 | 0,1869 | 1,0817 | 0,3224 | 0,9378 | 0,4491 | 0,738 | 0,0127 | 1,2891 | 0,0226 |
| Q8CJG5 | Gene | 3 | 0,9562 | 0,7992 | 0,6552 | 0,233 | 1,5338 | 0,0488 | 1,0942 | 0,6762 | 0,9227 | 0,7939 |
| P05370 | Glucose-6-phosphate 1-dehydrogenase | 16 | 1,0157 | 0,847 | 1,0285 | 0,7414 | 0,9947 | 0,9314 | 1,0827 | 0,1708 | 1,2136 | 0,0003 |
| Q6P6V0 | Glucose-6-phosphate isomerase | 13 | 0,859 | 0,1021 | 0,9669 | 0,5321 | 1,1179 | 0,0929 | 1,1489 | 0,1394 | 1,2446 | 0,0072 |
| P04797 | Glyceraldehyde-3-phosphate dehydrogenase | 28 | 1,0559 | 0,3012 | 1,2268 | 0,0077 | 0,8176 | 0,0404 | 1,0972 | 0,4988 | 0,8495 | 0,1048 |
| P56574 | Isocitrate dehydrogenase [NADP], mitochondrial | 3 | 1,0223 | 0,8926 | 1,0131 | 0,8828 | 0,8721 | 0,3389 | 1,3609 | 0,025 | 0,8545 | 0,1231 |
| B5DEN4 | L-lactate dehydrogenase | 14 | 0,7058 | 0,0001 | 1.146 | 0,00197 | 1.1828 | 0,0191 | 1.2617 | 0 | 1.416 | 0 |
| Q6P7A9 | Lysosomal alpha-glucosidase | 4 | 0,9754 | 0,7869 | 1,0694 | 0,6226 | 1,4268 | 0,048 | 0,8687 | 0,254 | 1 | 1 |
| Q6AYC4 | Macrophage-capping protein | 2 | 1,1578 | 0,3404 | 1,3646 | 0,0086 | 0,8334 | 0,049 | 0,7546 | 0,0948 | 1,3655 | 0,0617 |
| F1LP60 | Moesin (Fragment) | 40 | 1,0168 | 0,6741 | 0,8227 | 0,0003 | 1,4257 | 0 | 0,9895 | 0,8299 | 0,9461 | 0,1782 |
| P20070 | NADH-cytochrome b5 reductase 3 | 2 | 0,7339 | 0,0716 | 1,4911 | 0,0447 | 0,9088 | 0,5256 | 0,9743 | 0,903 | 0,8315 | 0,169 |
| Q6XD99 | Non-erythroid spectrin beta | 2 | 1,3375 | 0,0053 | 1,0624 | 0,4554 | 1,1109 | 0,3101 | 0,7568 | 0,0122 | 1,5134 | 0,0041 |
| P16617 | Phosphoglycerate kinase 1 | 29 | 0,775 | 0 | 1,0397 | 0,3579 | 1,0241 | 0,6829 | 1,1179 | 0,0446 | 1,3076 | 0 |
| P25113 | Phosphoglycerate mutase 1 | 9 | 0,7559 | 0,0052 | 1,1081 | 0,1211 | 1,1188 | 0,142 | 1,105 | 0,1848 | 1,3205 | 0,0024 |
| P54001 | Prolyl 4-hydroxylase subunit alpha-1 | 15 | 0,9432 | 0,3007 | 0,923 | 0,1417 | 1,26 | 0,0005 | 0,779 | 0,0002 | 0,9234 | 0,2253 |
| M0R9D5 | Protein Ahnak | 60 | 1,4134 | 0 | 0,9734 | 0,2788 | 1,2761 | 0 | 0,8082 | 0 | 2,1551 | 0 |
| D3ZIE9 | Protein Aldh18a1 | 5 | 1,2134 | 0,1403 | 1,038 | 0,7002 | 0,5751 | 0,0236 | 0,8643 | 0,2248 | 0,7301 | 0,0622 |
| M0R3 × 6 | Protein LOC100912203 | 6 | 0,8645 | 0,163 | 1,0325 | 0,6321 | 1,1276 | 0,2248 | 0,9918 | 0,9068 | 1,2992 | 0,0104 |
| D4A5L9 | Protein LOC679794 | 4 | 0,6761 | 0,0039 | 1,6157 | 0,0178 | 0,7517 | 0,0082 | 0,9865 | 0,8661 | 1,0755 | 0,375 |
| Q6P9U0 | Protein Serpinb6 | 8 | 0,9886 | 0,8679 | 1,0239 | 0,6691 | 1,3013 | 0,0066 | 0,8823 | 0,2774 | 1,1196 | 0,16 |
| D3ZF39 | Protein Uap1 | 10 | 1,0277 | 0,7709 | 1,0072 | 0,947 | 1,6544 | 0,0038 | 0,9744 | 0,7756 | 1,6238 | 0,0005 |
| B0BMT0 | RCG47746, isoform CRA_a | 90 | 1,0669 | 0,6161 | 0,7141 | 0,0061 | 1,4858 | 0,0047 | 0,5813 | 0,0012 | 0,7691 | 0,1418 |
| Q6IRL3 | Reticulon | 7 | 1,0111 | 0,8671 | 0,8818 | 0,3109 | 1,3224 | 0,0677 | 0,7406 | 0,0052 | 0,9133 | 0,4439 |
| B2GVB1 | S100 calcium binding protein A6 | 3 | 1,0605 | 0,5609 | 1,2045 | 0,2186 | 1,7118 | 0,0122 | 0,3918 | 0,0244 | 1,665 | 0,037 |
| Q5U3Z7 | Serine hydroxymethyltransferase | 3 | 0,7739 | 0,0051 | 0,9472 | 0,5152 | 1,0085 | 0,9511 | 1,1442 | 0,2328 | 1,0402 | 0,831 |
| F1M953 | Stress-70 protein, mitochondrial | 12 | 0,9377 | 0,2437 | 0,7924 | 0,0057 | 1,174 | 0,0025 | 0,8961 | 0,1199 | 1,5666 | 0 |
| P48500 | Triosephosphate isomerase | 16 | 0,6753 | 0,0004 | 1,174 | 0,0315 | 1,0854 | 0,3958 | 1,2639 | 0,1091 | 1,4017 | 0,0001 |
| Q9Z1A6 | Vigilin | 3 | 1,1784 | 0,0874 | 0,8426 | 0,0918 | 1,0244 | 0,862 | 0,9978 | 0,9727 | 1,2104 | 0,0345 |
| P81155 | Voltage-dependent anion-selective channel protein 2 | 6 | 1,0012 | 0,9931 | 1,0633 | 0,4458 | 1,2708 | 0,0343 | 0,8377 | 0,1616 | 0,992 | 0,9444 |
| Pluripotency | ||||||||||||
| P63102 | 14-3-3 protein zeta/delta | 24 | 0,9971 | 0,9573 | 0,9743 | 0,6297 | 1,0748 | 0,2065 | 0,8947 | 0,1104 | 1,1562 | 0,0250 |
| Q7TP91 | Ab1-205 | 3 | 0,9802 | 0,8434 | 1,2607 | 0,1387 | 0,9026 | 0,3795 | 1,0629 | 0,7975 | 0,6232 | 0,0302 |
| Q64640 | Adenosine kinase | 2 | 0,9609 | 0,8197 | 1,1207 | 0,3134 | 1,1155 | 0,346 | 1,0828 | 0,4536 | 0,6855 | 0,0454 |
| P39069 | Adenylate kinase isoenzyme 1 | 4 | 0,7781 | 0,0775 | 1,2606 | 0,053 | 1,0747 | 0,7679 | 0,9795 | 0,8759 | 1,6777 | 0,029 |
| P23928 | Alpha-crystallin B chain | 5 | 1,207 | 0,0573 | 1,8689 | 0,0119 | 2,2257 | 0,0003 | 0,2931 | 0,0035 | 1,5629 | 0,0723 |
| Q6IMZ3 | Annexin A6 | 24 | 1,2484 | 0,0001 | 1,1079 | 0,0199 | 0,9791 | 0,5623 | 1,0031 | 0,9354 | 1,0187 | 0,5863 |
| Q07936 | Annexin A2 | 22 | 1,2928 | 0 | 1,0234 | 0,6825 | 1,368 | 0,0001 | 0,8172 | 0,003 | 1,072 | 0,1567 |
| Q05175 | Brain acid soluble protein 1 | 3 | 0,9481 | 0,8617 | 1,209 | 0,454 | 1,3727 | 0,148 | 0,4584 | 0,0445 | 1,6462 | 0,1009 |
| Q6T487 | Brain-specific alpha actinin 1 isoform | 48 | 0,7786 | 0,0007 | 1,1584 | 0,0465 | 1,0249 | 0,6506 | 0,8259 | 0,0018 | 1,1052 | 0,2188 |
| Q8R4A2 | Caveolin 1 (Fragment) | 4 | 0,9676 | 0,8582 | 1,8465 | 0,0328 | 1,0661 | 0,7396 | 0,7326 | 0,1573 | 1,0726 | 0,6061 |
| P02454 | Collagen alpha-1(I) chain | 21 | 1,9314 | 0,0008 | 0,4642 | 0 | 1,3783 | 0 | 1,1251 | 0,0342 | 1,123 | 0,1023 |
| F1LS40 | Collagen alpha-2(I) chain | 19 | 1,486 | 0,0008 | 0,6971 | 0 | 1,1467 | 0,0083 | 0,9555 | 0,2908 | 1,3211 | 0,0003 |
| P07335 | Creatine kinase B-type | 3 | 0,7036 | 0,0109 | 1,1785 | 0,2497 | 1,4259 | 0,0546 | 0,8379 | 0,2386 | 1,2443 | 0,3604 |
| F1LMA7 | C-type mannose receptor 2 | 5 | 1,1259 | 0,191 | 0,8731 | 0,3184 | 0,8322 | 0,3836 | 0,8408 | 0,3689 | 1,7697 | 0,0009 |
| P47875 | Cysteine and glycine-rich protein 1 | 5 | 1,0995 | 0,2349 | 0,9264 | 0,4028 | 1,275 | 0,0227 | 0,7329 | 0,0396 | 1,2025 | 0,1773 |
| Q6AYI1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 9 | 0,9335 | 0,2865 | 0,9937 | 0,9257 | 0,8965 | 0,0659 | 1,2504 | 0,0032 | 0,9596 | 0,434 |
| Q62952 | Dihydropyrimidinase-related protein 3 | 8 | 1,2443 | 0,0364 | 1,5656 | 0,0028 | 0,858 | 0,2853 | 0,8247 | 0,0165 | 1,0088 | 0,9184 |
| Q4V8H8 | EH domain-containing protein 2 | 0 | 0,9656 | 0,7399 | 1,3396 | 0,2041 | 0,8004 | 0,248 | 1,6703 | 0,0471 | 0,7495 | 0,491 |
| Q68FR6 | Elongation factor 1-gamma | 9 | 1,0087 | 0,8562 | 0,9072 | 0,4157 | 1,2134 | 0,0011 | 0,9535 | 0,5667 | 0,8485 | 0,0109 |
| C0JPT7 | Filamin alpha | 100 | 1,2134 | 0 | 1,3022 | 0 | 0,8202 | 0 | 0,9147 | 0,0068 | 1,386 | 0 |
| D4A8D5 | Filamin, beta (Predicted) | 19 | 1,0951 | 0,0867 | 1,2541 | 0,0012 | 0,8193 | 0,0061 | 0,7937 | 0,0124 | 1,329 | 0,0009 |
| B6DYQ7 | Glutathione S-transferase pi | 4 | 1,0946 | 0,3206 | 1,1008 | 0,6055 | 1,5111 | 0,0241 | 3,1507 | 0,0004 | 0,3406 | 0,0033 |
| G3V913 | Heat shock 27kDa protein 1 | 5 | 1,6407 | 0,0562 | 0,9647 | 0,8462 | 1,5674 | 0,0118 | 0,636 | 0,038 | 1,3145 | 0,0077 |
| P63018 | Heat shock cognate 71 kDa protein | 30 | 0,9815 | 0,6625 | 1,2334 | 0,0221 | 0,8841 | 0,0143 | 1,0164 | 0,8526 | 1,1903 | 0,0184 |
| F1M3D3 | Heterogeneous nuclear ribonucleoprotein M | 3 | 0,6983 | 0,0017 | 1,0334 | 0,5966 | 0,9158 | 0,4502 | 1,1725 | 0,1239 | 1,0388 | 0,7714 |
| Q6IMY8 | Heterogeneous nuclear ribonucleoprotein U | 8 | 0,7965 | 0,0032 | 1,0228 | 0,7409 | 0,9994 | 0,9956 | 1,1924 | 0,0262 | 0,8534 | 0,1414 |
| P15865 | Histone H1.4 | 8 | 0,7411 | 0,0048 | 2,4013 | 0,0005 | 1,0711 | 0,2301 | 0,5016 | 0,0007 | 0,6377 | 0,007 |
| D3ZBN0 | Histone H1.5 | 4 | 1,911 | 0,0166 | 0,4907 | 0,0135 | 0,9905 | 0,9189 | 1,4031 | 0,0552 | 0,8838 | 0,2767 |
| G3V9C7 | Histone H2B | 20 | 1,2535 | 0,0051 | 0,9957 | 0,9764 | 0,8311 | 0,1792 | 1,4957 | 0,0005 | 0,7706 | 0,0027 |
| M0RBX6 | Histone H3 | 6 | 1,1323 | 0,058 | 1,4413 | 0,0125 | 1,201 | 0,0272 | 0,5991 | 0,0004 | 0,494 | 0,0002 |
| P62804 | Histone H4 | 13 | 1,4819 | 0,0057 | 0,5999 | 0,0004 | 1,1515 | 0,0641 | 1,7155 | 0,0008 | 0,6785 | 0,002 |
| Q6P6G9 | Hnrpa1 protein | 8 | 0,6091 | 0,0304 | 0,9626 | 0,6587 | 0,943 | 0,663 | 1,1142 | 0,7189 | 0,969 | 0,7865 |
| P50503 | Hsc70-interacting protein | 4 | 0,9785 | 0,7614 | 1,0509 | 0,7519 | 1,1426 | 0,4415 | 0,8392 | 0,2581 | 1,5699 | 0,0074 |
| P49134 | Integrin beta-1 | 6 | 1,5471 | 0,0002 | 0,9023 | 0,1179 | 1,5398 | 0,0098 | 0,7217 | 0,0064 | 1,311 | 0,0037 |
| G3V7Q7 | IQ motif containing GTPase activating protein 1 (Predicted), isoform CRA_b | 29 | 0,9058 | 0,0331 | 0,8665 | 0,0016 | 1,2223 | 0 | 1,0398 | 0,2744 | 0,8961 | 0,0032 |
| Q6TXE9 | LRRGT00050 | 4 | 0,8206 | 0,0425 | 0,8878 | 0,4999 | 1,0972 | 0,6832 | 1,5 | 0,0036 | 0,8044 | 0,129 |
| Q6TUD1 | LRRGT00113 | 2 | 0,7362 | 0,0289 | 1,0159 | 0,9238 | 1,0387 | 0,7991 | 1,0836 | 0,5779 | 0,9606 | 0,8364 |
| Q5M7W5 | Microtubule-associated protein 4 | 2 | 1,6111 | 0,1601 | 0,845 | 0,5023 | 1,0907 | 0,433 | 0,6693 | 0,1236 | 1,6243 | 0,0414 |
| B2GV99 | Myl6 protein | 11 | 1,0049 | 0,9535 | 1,1269 | 0,1948 | 1,0655 | 0,29 | 0,9193 | 0,1676 | 1,3394 | 0,0011 |
| G3V9Y1 | Myosin, heavy polypeptide 10, non-muscle, isoform CRA_b | 51 | 0,9356 | 0,0771 | 1,0793 | 0,1827 | 0,8786 | 0,0186 | 0,8988 | 0,0205 | 0,792 | 0,0018 |
| G3V6P7 | Myosin, heavy polypeptide 9, non-muscle | 98 | 0,9405 | 0,0071 | 1,1877 | 0 | 1,0117 | 0,6464 | 0,957 | 0,2338 | 1,3299 | 0 |
| P05982 | NAD(P)H dehydrogenase [quinone] 1 | 8 | 1,3454 | 0,0036 | 0,7746 | 0,0108 | 1,2457 | 0,0611 | 0,817 | 0,2198 | 1,7303 | 0,0017 |
| G3V8R1 | Nucleobindin 2, isoform CRA_b | 3 | 0,7234 | 0,0259 | 2,0743 | 0,0023 | 0,5945 | 0,0061 | 0,8133 | 0,1039 | 1,5101 | 0,0176 |
| F1M4W3 | Palladin (Fragment) | 6 | 1,0033 | 0,9633 | 0,8726 | 0,1096 | 1,0418 | 0,6509 | 0,6823 | 0,0117 | 1,0929 | 0,2707 |
| P52944 | PDZ and LIM domain protein 1 | 8 | 1,0741 | 0,3026 | 1,0802 | 0,1599 | 1,3743 | 0,0005 | 0,9648 | 0,7456 | 1,2349 | 0,03 |
| Q62920 | PDZ and LIM domain protein 5 | 17 | 0,9467 | 0,5156 | 0,6947 | 0,0022 | 1,4784 | 0,0057 | 0,6885 | 0,0358 | 0,8014 | 0,071 |
| Q6AYQ9 | Peptidyl-prolyl cis-trans isomerase | 6 | 0,9218 | 0,2069 | 0,9782 | 0,6989 | 1,2033 | 0,0649 | 0,7408 | 0,0209 | 0,837 | 0,0475 |
| Q62658 | Peptidyl-prolyl cis-trans isomerase FKBP1A | 2 | 1,254 | 0,0638 | 1,1087 | 0,2151 | 1,0291 | 0,6981 | 0,8516 | 0,0786 | 1,38 | 0,0149 |
| D3ZAF5 | Periostin, osteoblast specific factor (Predicted), isoform CRA_a | 4 | 0,5315 | 0,1266 | 1,4489 | 0,0583 | 0,7663 | 0,0352 | 1,3251 | 0,0507 | 0,8907 | 0,4068 |
| Q63716 | Peroxiredoxin-1 | 13 | 0,8935 | 0,0404 | 1,0622 | 0,5224 | 1,083 | 0,5586 | 0,884 | 0,3841 | 1,2919 | 0,0335 |
| P35704 | Peroxiredoxin-2 | 5 | 0,9438 | 0,6729 | 1,3399 | 0,0331 | 0,9268 | 0,3898 | 0,8375 | 0,2252 | 1,1003 | 0,4352 |
| Q9R063 | Peroxiredoxin-5, mitochondrial | 5 | 1,0571 | 0,6808 | 0,7826 | 0,0577 | 1,2214 | 0,0908 | 0,8798 | 0,195 | 1,4277 | 0,0498 |
| F1LPK7 | Phospholipid scramblase 3 | 5 | 1,3539 | 0,015 | 0,7564 | 0,0206 | 1,2564 | 0,0285 | 0,9783 | 0,8008 | 1,0061 | 0,9704 |
| G3V8L9 | Polymerase I and transcript release factor | 10 | 1,0181 | 0,7755 | 1,5741 | 0,0001 | 1,2442 | 0,0026 | 0,6574 | 0,0001 | 1,2032 | 0,1746 |
| G3V9I0 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | 15 | 0,6772 | 0,0001 | 1,3393 | 0,0139 | 0,8435 | 0,0459 | 1,1106 | 0,2824 | 1,033 | 0,765 |
| D3ZRX9 | Protein Cnn2 | 9 | 0,9803 | 0,7232 | 0,9782 | 0,6955 | 1,1542 | 0,0382 | 0,7727 | 0,0037 | 1,1053 | 0,1116 |
| G3V6T7 | Protein disulfide isomerase associated 4 | 4 | 1,094 | 0,3778 | 1,5291 | 0,0042 | 0,7583 | 0,087 | 0,8527 | 0,0368 | 1,1596 | 0,0466 |
| P04785 | Protein disulfide-isomerase | 18 | 0,9524 | 0,2161 | 0,9019 | 0,0205 | 1,1449 | 0,002 | 0,91 | 0,0672 | 1,3562 | 0 |
| P11598 | Protein disulfide-isomerase A3 | 23 | 1,0044 | 0,9331 | 1,1813 | 0,0003 | 1,0096 | 0,899 | 0,888 | 0,1863 | 1,151 | 0,1114 |
| Q63081 | Protein disulfide-isomerase A6 | 9 | 0,7832 | 0,0043 | 1,1044 | 0,262 | 1,2788 | 0,0234 | 0,9335 | 0,4195 | 1,0394 | 0,5727 |
| D3ZHA0 | Protein Flnc | 28 | 0,9537 | 0,2739 | 1,6131 | 0 | 0,8375 | 0,0128 | 0,9354 | 0,1802 | 1,1796 | 0,0125 |
| E2RUH2 | Protein LOC100360501 | 3 | 0,8715 | 0,4837 | 1,2547 | 0,0379 | 0,7763 | 0,024 | 1,2418 | 0,1487 | 0,7978 | 0,0359 |
| M0R7B4 | Protein LOC684828 | 6 | 1,9171 | 0,003 | 0,4959 | 0,0035 | 1,0813 | 0,3542 | 1,3227 | 0,0305 | 0,8447 | 0,1021 |
| F1MA29 | Protein LOC685520 | 5 | 0,7506 | 0,0026 | 1,15 | 0,0892 | 0,9181 | 0,414 | 1,0561 | 0,438 | 1,066 | 0,4416 |
| D3ZUB0 | Protein Rcn1 | 2 | 1,0185 | 0,8167 | 0,8941 | 0,2217 | 1,1262 | 0,2053 | 0,8607 | 0,297 | 1,3273 | 0,0313 |
| I6L9G5 | Protein Rcn3 | 2 | 1,0873 | 0,4834 | 0,5646 | 0,023 | 1,147 | 0,2936 | 0,9716 | 0,7715 | 1,3833 | 0,2381 |
| D4A1P2 | Protein Rpl10l | 7 | 1,0101 | 0,8587 | 0,8912 | 0,0814 | 0,8855 | 0,0554 | 1,4153 | 0,0002 | 0,6894 | 0,0001 |
| F1M853 | Protein Rrbp1 | 12 | 0,9865 | 0,8266 | 0,9487 | 0,2254 | 1,4058 | 0,0002 | 0,6416 | 0,0002 | 1,4396 | 0 |
| P05942 | Protein S100-A4 | 8 | 1,3344 | 0,0883 | 0,8596 | 0,3432 | 2,371 | 0,0005 | 0,6816 | 0,0449 | 1,6256 | 0,0034 |
| B0BMT9 | Protein Sqrdl | 5 | 0,8772 | 0,1123 | 0,8745 | 0,2138 | 1,3482 | 0,0318 | 0,672 | 0,0272 | 1,1637 | 0,2917 |
| P50399 | Rab GDP dissociation inhibitor beta | 5 | 0,6527 | 0 | 1,1226 | 0,1842 | 0,8087 | 0,0191 | 1,2167 | 0,0909 | 0,8327 | 0,0497 |
| Q5FVG5 | Similar to tropomyosin 1, embryonic fibroblast-rat, isoform CRA_c | 21 | 0,8189 | 0,0635 | 0,8236 | 0,0664 | 1,5332 | 0,0055 | 0,4284 | 0,0029 | 0,9622 | 0,622 |
| Q6IRH6 | Slc25a3 protein | 5 | 0,6221 | 0,0046 | 1,2418 | 0,0813 | 0,9018 | 0,3869 | 1,2937 | 0,0271 | 0,7334 | 0,0095 |
| P06685 | Sodium/potassium-transporting ATPase subunit alpha-1 | 6 | 1,0612 | 0,5229 | 0,847 | 0,0206 | 0,9521 | 0,6531 | 1,1204 | 0,4092 | 0,8973 | 0,1495 |
| P16975 | SPARC | 5 | 1,2574 | 0,0585 | 0,8963 | 0,1879 | 1,0501 | 0,5303 | 0,9358 | 0,5629 | 1,2361 | 0,0437 |
| Q63413 | Spliceosome RNA helicase Ddx39b | 4 | 0,7567 | 0,0206 | 1,0276 | 0,7991 | 0,904 | 0,431 | 1,3219 | 0,0189 | 0,679 | 0,0523 |
| Q6IRK8 | Spna2 protein | 9 | 1,3867 | 0 | 1,211 | 0,003 | 1,0232 | 0,7136 | 0,7877 | 0,0016 | 1,7205 | 0 |
| D4A8Y5 | Staphylococcal nuclease domain-containing protein 1 | 3 | 0,919 | 0,451 | 1,2908 | 0,0445 | 0,8256 | 0,0815 | 1,2376 | 0,0644 | 0,7802 | 0,1202 |
| Q71SA3 | Thrombospondin 1 | 7 | 0,8058 | 0,0398 | 0,7166 | 0,0007 | 1,3192 | 0,0007 | 0,9713 | 0,6767 | 1,4974 | 0,0023 |
| P31232 | Transgelin | 39 | 1,2096 | 0,0003 | 1,2967 | 0,0743 | 1,133 | 0,0136 | 0,5637 | 0,0001 | 1,7372 | 0 |
| Q5XFX0 | Transgelin-2 | 17 | 0,9888 | 0,8414 | 1,0039 | 0,955 | 1,3666 | 0,0009 | 0,9786 | 0,7678 | 1,1396 | 0,048 |
| Q6AYT3 | tRNA-splicing ligase RtcB homolog | 4 | 0,6896 | 0,019 | 0,9867 | 0,8815 | 0,8815 | 0,1894 | 1,1372 | 0,1808 | 0,8525 | 0,4565 |
| Q63610 | Tropomyosin alpha-3 chain | 9 | 0,9843 | 0,838 | 1,4987 | 0,0199 | 1,2255 | 0,0709 | 0,6776 | 0,0538 | 1,8819 | 0,0153 |
| P09495 | Tropomyosin alpha-4 chain | 12 | 0,9739 | 0,7802 | 1,356 | 0,0682 | 1,0117 | 0,925 | 0,8335 | 0,1905 | 1,5771 | 0,0401 |
| G3V6C4 | UDP-glucose 6-dehydrogenase | 8 | 1,0597 | 0,478 | 0,9444 | 0,6809 | 1,256 | 0,011 | 1,0009 | 0,9911 | 1,4008 | 0,0135 |
| Q63355 | Unconventional myosin-Ic | 10 | 1,2333 | 0,0007 | 0,8095 | 0,0071 | 1,2252 | 0,0431 | 1,0652 | 0,3451 | 0,8854 | 0,0253 |
| P31000 | Vimentin | 110 | 1,0703 | 0,0545 | 1,0991 | 0,0394 | 1,1756 | 0,0018 | 0,76 | 0 | 0,9955 | 0,9047 |
| Proliferation | ||||||||||||
| P62268 | 40S ribosomal protein S23 | 3 | 1,0729 | 0,3955 | 0,7949 | 0,0312 | 1,0116 | 0,8769 | 0,9447 | 0,6609 | 1,0981 | 0,4568 |
| M0RD75 | 40S ribosomal protein S6 (Fragment) | 5 | 1,213 | 0,0348 | 0,7478 | 0,0343 | 1,0648 | 0,6105 | 0,8839 | 0,3222 | 1,2172 | 0,0637 |
| B2RYR8 | 40S ribosomal protein S8 | 5 | 1,156 | 0,0743 | 0,7651 | 0,0113 | 0,9563 | 0,6408 | 1,1034 | 0,3318 | 1,0474 | 0,6037 |
| P29314 | 40S ribosomal protein S9 | 10 | 1,2232 | 0,0078 | 0,7297 | 0,0002 | 1,1604 | 0,014 | 1,0442 | 0,4378 | 1,0231 | 0,6427 |
| P38983 | 40S ribosomal protein SA | 7 | 1,0045 | 0,948 | 0,8267 | 0,0864 | 1,0707 | 0,3666 | 1,3083 | 0,0159 | 0,8177 | 0,0379 |
| P63039 | 60 kDa heat shock protein, mitochondrial | 14 | 1,0284 | 0,7397 | 1,0424 | 0,6331 | 1,0209 | 0,827 | 0,7502 | 0,0126 | 1,1048 | 0,3793 |
| Q6PDV7 | 60S ribosomal protein L10 | 8 | 1,2944 | 0,0232 | 0,7476 | 0,0503 | 1,0811 | 0,4911 | 0,9186 | 0,3735 | 1,079 | 0,2893 |
| P41123 | 60S ribosomal protein L13 | 4 | 1,2788 | 0,1225 | 0,8351 | 0,0607 | 1,0613 | 0,7175 | 1,269 | 0,0179 | 0,8472 | 0,0781 |
| P61314 | 60S ribosomal protein L15 | 2 | 1,0608 | 0,6431 | 0,9332 | 0,5211 | 0,8939 | 0,228 | 1,6856 | 0,0065 | 0,7429 | 0,0338 |
| Q0QEW8 | 60S ribosomal protein L18 (Fragment) | 3 | 0,8899 | 0,3903 | 1,0505 | 0,6391 | 0,9184 | 0,5621 | 1,4963 | 0,0365 | 0,7225 | 0,0735 |
| P62718 | 60S ribosomal protein L18a | 4 | 1,0118 | 0,8521 | 0,8707 | 0,2832 | 1,0157 | 0,9288 | 1,3147 | 0,032 | 0,8694 | 0,1711 |
| P62832 | 60S ribosomal protein L23 | 6 | 1,2256 | 0,0212 | 0,838 | 0,0334 | 0,9969 | 0,9615 | 0,9723 | 0,8064 | 1,1553 | 0,1309 |
| P83732 | 60S ribosomal protein L24 | 7 | 1,5524 | 0,0024 | 0,5064 | 0,0004 | 1,2108 | 0,0391 | 0,8698 | 0,2155 | 1,2834 | 0,0175 |
| P25886 | 60S ribosomal protein L29 | 3 | 1,5331 | 0,0443 | 0,9651 | 0,7362 | 1,0984 | 0,4167 | 0,8404 | 0,5923 | 0,7419 | 0,2733 |
| P21531 | 60S ribosomal protein L3 | 5 | 1,0552 | 0,6436 | 0,8494 | 0,0879 | 0,932 | 0,5476 | 1,463 | 0,0213 | 0,7223 | 0,0148 |
| Q6P3V9 | 60S ribosomal protein L4 | 9 | 1,419 | 0,0021 | 0,627 | 0,0002 | 1,0227 | 0,7849 | 0,9688 | 0,6057 | 1,139 | 0,0543 |
| P09895 | 60S ribosomal protein L5 | 6 | 0,9424 | 0,3628 | 0,9732 | 0,6933 | 0,9466 | 0,5073 | 1,2345 | 0,0225 | 0,8597 | 0,1305 |
| H7C5Y5 | 60S ribosomal protein L6 | 7 | 1,3496 | 0,0261 | 0,6716 | 0,0068 | 1,0461 | 0,66 | 1,0542 | 0,451 | 1,0501 | 0,6358 |
| Q6P790 | 60S ribosomal protein L6 (Fragment) | 7 | 1,2191 | 0,0025 | 1,0302 | 0,7281 | 1,0286 | 0,6572 | 0,8965 | 0,2512 | 0,987 | 0,8763 |
| P05426 | 60S ribosomal protein L7 | 5 | 1,3498 | 0,0462 | 0,6788 | 0,065 | 1,1405 | 0,3092 | 0,9309 | 0,3885 | 1,0532 | 0,5387 |
| P85970 | Actin-related protein 2/3 complex subunit 2 | 11 | 0,9564 | 0,6557 | 0,9226 | 0,2715 | 1,3327 | 0,0061 | 1,0044 | 0,938 | 1,0774 | 0,3839 |
| Q9Z1P2 | Alpha-actinin-1 | 77 | 0,8727 | 0,0028 | 0,8694 | 0,0002 | 1,3424 | 0,0013 | 0,8164 | 0,0001 | 0,9543 | 0,1358 |
| Q9QXQ0 | Alpha-actinin-4 | 50 | 1,2074 | 0,0004 | 1,1204 | 0,037 | 1,0349 | 0,3567 | 0,9069 | 0,2105 | 1,1419 | 0,036 |
| Q66HH8 | Annexin 5 | 9 | 0,9925 | 0,8998 | 1,0383 | 0,7603 | 0,9707 | 0,811 | 0,9909 | 0,8915 | 1,4149 | 0,0034 |
| P45592 | Cofilin-1 | 12 | 1,2442 | 0,0041 | 1,0603 | 0,5475 | 1,0804 | 0,2025 | 0,9203 | 0,5481 | 1,2685 | 0,011 |
| D3ZH41 | Cytoskeleton-associated protein 4 (Predicted) | 12 | 0,7853 | 0,0004 | 1,107 | 0,0513 | 0,9476 | 0,3458 | 0,7919 | 0,0005 | 1,2344 | 0,0033 |
| Q6AYH5 | Dynactin subunit 2 | 3 | 1,151 | 0,2739 | 0,8845 | 0,3112 | 1,1661 | 0,2508 | 0,8114 | 0,3778 | 1,3787 | 0,0248 |
| P52555 | Endoplasmic reticulum resident protein 29 | 2 | 1,0186 | 0,8459 | 1,3064 | 0,1141 | 1,2519 | 0,2488 | 0,8626 | 0,2226 | 1,5742 | 0,0319 |
| Q6P3V8 | Eukaryotic translation initiation factor 4A1 | 13 | 1,0465 | 0,4461 | 0,8673 | 0,0328 | 1,055 | 0,416 | 1,0269 | 0,6429 | 0,7893 | 0,0001 |
| P04937 | Fibronectin | 17 | 1,1267 | 0,3104 | 0,7887 | 0,0119 | 1,2538 | 0,0001 | 0,6481 | 0,0907 | 2,4985 | 0,0056 |
| Q6P792 | Four and a half LIM domains 1 | 6 | 0,7431 | 0,0006 | 1,7903 | 0,0001 | 1,1108 | 0,096 | 0,7768 | 0,0085 | 0,8731 | 0,0375 |
| P11762 | Galectin-1 OS=Rattus norvegicus | 14 | 0,7674 | 0,0204 | 0,7313 | 0,053 | 1,2804 | 0,0564 | 0,8213 | 0,4313 | 1,1116 | 0,227 |
| B6DYQ2 | Glutathione S-transferase mu 2 | 5 | 1,1023 | 0,4484 | 1,0507 | 0,6202 | 0,8053 | 0,0678 | 0,8616 | 0,3082 | 1,2838 | 0,0235 |
| P63245 | Guanine nucleotide-binding protein subunit beta-2-like 1 | 6 | 1,0188 | 0,7481 | 0,8543 | 0,044 | 1,0255 | 0,6962 | 1,0046 | 0,963 | 0,9843 | 0,8938 |
| Q6P7Q4 | Lactoylglutathione lyase | 6 | 0,8942 | 0,2376 | 0,9113 | 0,2336 | 1,2099 | 0,0449 | 0,9186 | 0,2639 | 1,0132 | 0,8509 |
| G3V8L3 | Lamin A, isoform CRA_b | 26 | 0,9048 | 0,0029 | 0,968 | 0,4478 | 1,1812 | 0,0001 | 1,0126 | 0,746 | 1,2067 | 0 |
| Q99MZ8 | LIM and SH3 domain protein 1 | 5 | 1,4773 | 0,0032 | 1,0103 | 0,9316 | 1,1251 | 0,1144 | 0,7622 | 0,0071 | 1,3922 | 0,0031 |
| O08557 | N(G), N(G)-dimethylarginine dimethylaminohydrolase 1 | 4 | 0,8682 | 0,4556 | 2,2145 | 0,0173 | 0,5358 | 0,0395 | 0,7504 | 0,1763 | 1,2909 | 0,0453 |
| Q6S3A0 | Plectin 6 | 28 | 1,0437 | 0,2665 | 0,9378 | 0,1139 | 1,1564 | 0,0026 | 0,996 | 0,9259 | 1,2269 | 0,0027 |
| D4A4Z9 | Protein Ktn1 | 7 | 0,9773 | 0,8388 | 0,901 | 0,2289 | 1,3423 | 0,0487 | 0,7373 | 0,0131 | 1,0952 | 0,3487 |
| D3ZPL5 | Protein LOC100361311 | 10 | 1,2843 | 0,0356 | 0,683 | 0,017 | 1,0148 | 0,8697 | 0,9663 | 0,7274 | 1,1144 | 0,0623 |
| M0RCY2 | Protein LOC683961 | 6 | 0,9886 | 0,9133 | 0,9596 | 0,6555 | 0,761 | 0,0353 | 1,4335 | 0,0202 | 0,591 | 0,0003 |
| D3ZN21 | Protein RGD1309586 | 6 | 0,951 | 0,6075 | 0,9746 | 0,7735 | 0,957 | 0,6381 | 1,2502 | 0,0226 | 0,8539 | 0,0205 |
| D4A6W6 | Protein RGD1561333 | 6 | 1,7423 | 0,0038 | 0,5049 | 0,0038 | 1,2265 | 0,2464 | 0,8945 | 0,5405 | 1,1717 | 0,2285 |
| D4A6W6 | Protein RGD1561333 | 5 | 1,1352 | 0,1387 | 1,0601 | 0,5478 | 0,9173 | 0,2914 | 1,5332 | 0,0034 | 0,6202 | 0,0124 |
| F1LT35 | Protein RGD1564606 (Fragment) | 6 | 1,1172 | 0,1379 | 1,1037 | 0,4799 | 1,0681 | 0,4407 | 0,772 | 0,0124 | 1,2544 | 0,0157 |
| G3V852 | Protein Tln1 | 38 | 1,2686 | 0 | 1,2229 | 0 | 0,8272 | 0 | 0,9642 | 0,3307 | 1,3768 | 0 |
| Q4QQV0 | Protein Tubb6 | 22 | 1,0776 | 0,4062 | 0,8668 | 0,3417 | 1,2995 | 0,0298 | 0,946 | 0,6332 | 1,0638 | 0,6569 |
| Q6P3E1 | Rps16 protein (Fragment) | 7 | 1,6881 | 0,0047 | 0,5471 | 0,0061 | 1,508 | 0,0481 | 0,8378 | 0,1083 | 1,2384 | 0,0629 |
| Q9QZR6 | Septin-9 | 6 | 1,0595 | 0,4348 | 0,9334 | 0,3019 | 1,2185 | 0,0277 | 0,8772 | 0,1483 | 1,017 | 0,8438 |
| Q6LDS4 | Superoxide dismutase [Cu-Zn] | 6 | 1,1464 | 0,0737 | 1,169 | 0,0483 | 0,8679 | 0,0643 | 1,0068 | 0,9549 | 1,3603 | 0,0094 |
| P07895 | Superoxide dismutase [Mn], mitochondrial | 10 | 0,76 | 0,2573 | 0,9976 | 0,9829 | 2,2825 | 0,02 | 1,074 | 0,2946 | 1,3438 | 0,0046 |
| P28480 | T-complex protein 1 subunit alpha | 6 | 0,7782 | 0,0579 | 1,5251 | 0,0086 | 0,9731 | 0,7177 | 1,2381 | 0,2501 | 0,9675 | 0,8659 |
| Q68FQ0 | T-complex protein 1 subunit epsilon | 4 | 0,8933 | 0,2273 | 1,0047 | 0,9576 | 0,9601 | 0,7021 | 1,1826 | 0,1497 | 0,8175 | 0,0229 |
| Q6P502 | T-complex protein 1 subunit gamma | 5 | 1,0456 | 0,4189 | 0,8549 | 0,0333 | 1,0502 | 0,74 | 1,0936 | 0,3134 | 0,9009 | 0,3475 |
| P11232 | Thioredoxin | 9 | 0,9519 | 0,7004 | 0,9806 | 0,8828 | 1,1098 | 0,4942 | 0,9058 | 0,4438 | 1,2619 | 0,0373 |
| Q99PD6 | Transforming growth factor beta-1-induced transcript 1 protein | 6 | 1,0858 | 0,7029 | 0,7535 | 0,8138 | 1,2703 | 0,2603 | 0,635 | 0,0115 | 0,8607 | 0,8969 |
| P68370 | Tubulin alpha-1A chain OS=Rattus norvegicus GN=Tuba1a PE=1 SV=1 | 19 | 1,1996 | 0,0477 | 1,1154 | 0,0721 | 0,7776 | 0,0946 | 0,9411 | 0,7092 | 0,7696 | 0,0004 |
| R9PXU6 | Vinculin | 57 | 1,1908 | 0 | 0,9981 | 0,9534 | 1,4169 | 0 | 0,8383 | 0,0001 | 1,1453 | 0 |
Figure 3. Validation of iTRAQ analysis.
(A) Significant activates pathways obtained by comparing modulated proteins obtained by iTRAQ analysis employing functional annotations according to the String 9.0 software. Small numbers on the right of each bar are the modulated protein involved in each process. (B) Western blot analysis of Lamin A/C, Vimentin and Superoxide dismutase 2 (SOD-2). Tubulin was used as housekeeping. On the left molecular weight of each protein is shown. On the bottom the group’s source of mesenchymal stem cells used. The gels have been run under the same experimental conditions. (C) Vinculin gene expression using real-time reverse transcriptase PCR (qRT-PCR) analysis normalized by expression of HPRT gene used as housekeeping.
Figure 4. Metabolism profile from rats mesenchymal stem cells at different age.
(A) 2′,7′-dichlorofluorescein signal measured by flow cytometry to check ROS intracellular (B) Mitosox signal measured by flow cytometry to check ROS mitochondrial. (C) Lactate-dehydrogenase (LDH) activity measured by spectrophotometer analysis. (D) Pentose phosphate pathway activity measured by spectrometer analysis. G6PDH = Glucose-6-phosphate 1-dehydrogenase; 6GPDH = 6-phosphogluconate-dehydrogenase. *p value less than 0.01 compared with previous group and #p value less than 0.05 compared with previous group, were considered statistically significant using Mann-Whitney-U tests.
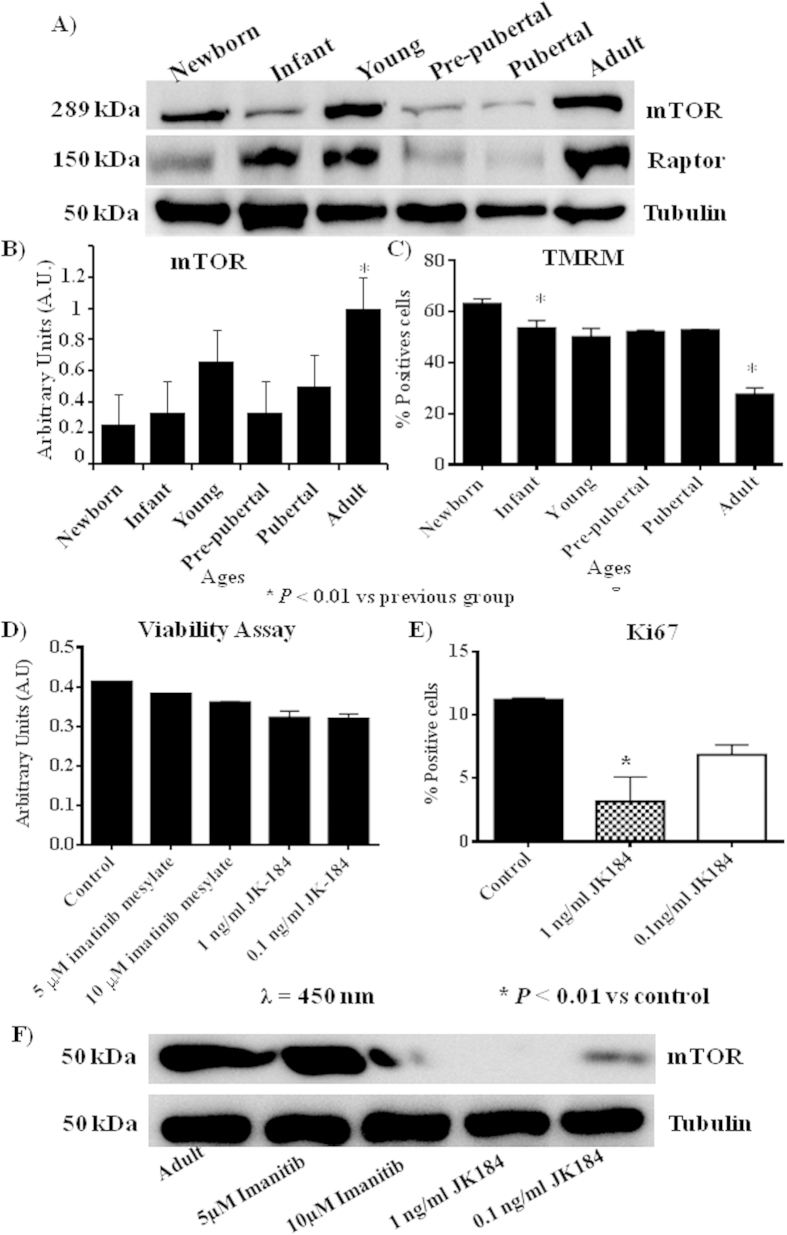
Figure 5. mTOR pathway profile from rat mesenchymal stem cells at different age (A) Western blot of mTOR pathway, mTOR and raptor, Tubulin was used as housekeeping.
(B) Densitometry analysis of westerns of mTOR normalized with respect to Tubulin. *p value less than 0.01 compared with previous group. (C) Tetramethylrhodamine, methyl ester (TMRM) dye accumulated in active mitochondria with intact potentials, was used to detect functional mitochondria in the MSCs at different ages following functional mitochondrial staining protocol from commercial. (D) Viability assay of mesenchymal stem cells from adult group incubated with 10 μM or 5 μM of imatinib mesylate or 1 ng/ml or 0.1 ng/ml of JK184. (E) Flow cytometry of Ki67 from mesenchymal stem cells of adult group incubated with 1 ng/ml or 0.1 ng/ml of JK184 in the medium. F) Western blot of mTOR and Tubulin of mesenchymal stem cells from adult group after incubating with 10 μM or 5 μM of imatinib mesylate or 1 ng/ml or 0.1 ng/ml of JK184 in the medium. Control = mesenchymal stem cells incubated with growth medium alone. The gels have been run under the same experimental conditions. *p value less than 0.01 compared with control group was considered statistically significant using Mann-Whitney-U tests.
Immunoblot analysis indicated that mTOR and Raptor were statistically significant (p < 0.05) lower in pre-pubertal and pubertal groups with respect to the other groups studied. Adult group presented the statistically significant (p < 0.05) most increased level of mTOR and Raptor (Fig. 5A), the permanent dye TMRM analysis indicated a decrease in functional mitochondria which was statistically significant (p < 0.01) in infant and adult groups with respect to newborn and pubertal groups respectively (Fig. 5B). Viability assay using two physiological concentrations of Imatinib mesylate or JK184 did not affect the cells in culture (Fig. 5C). JK184 decreased statistically significant (p < 0.01) the expression of KI67 at 1mg/ml dose in culture (Fig. 5D). mTOR decreased dramatically in adult group when the cells were incubated with Imatinib mesylate or JK184 (Fig. 5E).
Discussion
It is good known that long-term in vitro culture, but not chronological aging, alters their morphology, susceptibility to senescence and mitochondrial function. Thus, independent from donor animal age, in vitro aging of MSCs seems to result in complete loss of their progenitor characteristics9. Accordingly, functional analysis demonstrated altered mitochondrial morphology, decreased antioxidant capacities and elevated ROS levels in long-term cultivated independently the aged of the donor. Our present study is limited to the usage of rat MSCs instead can be able to apply on human MSCs; it provides direct comparison between chronological aged MSC not only at the cellular but also at the molecular level.
MSCs populations from different ageing groups were characterized by flow cytometry to check percentage of positive cells for mesenchymal markers, CD29 and CD90, and to check that were negative for hematopoietic markers (CD34, CD45) (Fig. 1A). We did not observe statistical significant differences between the mesenchymal markers into the different MSCs aging group studied (Fig. 2C). These were coincident with results published by Jin et al.6 indicating that MSCs have similar levels of surface antigen expression included MSCs from different tissues. Although the mesenchymal markers were not as abundant as published by Harting et al.22 these cells were able to adhere to the plastic plate and this is an intrinsic characteristic of mesenchymal stem cells and all of the groups were able to differentiate towards several mesoderm lineages (Fig. 2A,B). iTRAQ analysis is an adequate technique to study complex samples like we have used in this work23. Our results by iTRAQ analysis allowed the identification of 1.072 proteins. 201 of them were statistically significant modulated between groups (Table 2). Our study represents a step further from a previous iTRAQ-based study24 where 156 differentially expressed proteins were detected. Furthermore we compared six chronologically different groups, giving more strength to our study. To generate the quantitative proteome using iTRAQ labeling, it was first determined the labeling efficiency, which exceeded 99% (data not shown). Next, the cut-off for significant fold-change was determined based on the 2 biological replicates of two iTRAQ experiments which were chosen based on the following criteria: it contained more than 3 unique peptides (>95%) and p value < 0.05 for the 114/119 reporter ions. Accordingly, 90% of the commonly observed in the biological replicates fell within 25% of the respective experimental variation (Data don´t shown). The fold-change thresholds of >1.20 or <0.80 was set to identify true differences between expression of reporter ions.
One of the aims of our study was to establish the differences into proliferation process relating them to chronological donor age. Our results indicated that chronological age is directly influencing the expression of proliferation marker Ki6725 because of the lowest levels of Ki67 corresponded with less cells number in proliferation assays in infant and pre-pubertal groups. On the other way high levels of CD117, a self-renewal marker in MSCs as indicated Blazquez-Martinez et al.26 were corresponding to higher cells number in proliferation assay of pubertal and adult groups of animal respect to the rest groups (Fig. 1B,C).
60 modulated proteins found in our iTRAQ analysis were involved in proliferation as 60S ribosomal proteins with different sedimentation speed like 60S RP L10, 60S RP L9, 60S RP L23, 60S RP L24, 60S RP L4, 60S RP L6 and 60S RP L7 were over-expressed in young group respect to the others indicating their increased potential of pluripotency which would be in concordance with its low expression of Nanog gene by RT-PCR analysis (Fig. 2C) in this group as also Das et al.27 found over-expressed these proteins in different animal model process. Our results indicated that expression of Vinculin gene (Fig. 3C) was very low in newborn and young which were the most proliferative groups (Fig. 1B) on the opposite way pre-pubertal and adult presented high Vinculin gene expression coincidently with less proliferative potential. Toma-Jonik et al.28 published very recently that Vinculin, which is a protein involved in cell motility and adherence, was down-regulated in cells with great mobilization and proliferation potential like melanoma cells and at the same time Piltti et al.29 published that Rho kinase inhibitor (ROCKi) treatment increased the cellular proliferation up, in human foreskin fibroblast cells and, significantly less Vinculin-associated focal adhesions were present in these ROCKi-treated cells30. 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1), is an enzyme which generates glucocorticoids in intact cells, was found in our iTRAQ analysis significant increased in prepubertal group, this fact could explain the decrease proliferation potential and increase adypogenic differentiation in prepubertal group. All this is coincident with Bujalska et al.31 who published that 11β-HSD1 activity is uncommitted adipose stromal cells may facilitate proliferation rather than differentiation. Transforming growth factor β1 (TGF-β1) induces senescence in BM-MSCs via increases the mitochondrial reactive oxygen species production and also the ROS intracellular production is associated with decreasing mitochondrial membrane potential, DNA damage and cell senescence32,33, this fact could explain that statistically significant (p < 0.05) decreasing of total ROS in pre-pubertal group because TGF-β1 was found statistically significant (p < 0.05) low with respect to other groups in the iTRAQ analysis.
86 modulated proteins found in our iTRAQ analysis were involved in pluripotency process. Terme et al.34 showed that pluripotent cells had decreased levels of H1.0 and increased levels of H1.1, H1.3, and H1.5 compared with differentiated cells. Our results also indicated that H4 was statistically significant decrease in adult group which could point towards their less pluripotency with respect to the other groups, differentiation of embryonic stem cells is accompanied by a global reduction of panacetylation of histones H3 and H4 suggesting that histone acetylation plays an important role in maintenance of embryonic stem cells pluripotency35. Results published by Bermeo et al.36 indicate that MSCs over-expressing Lamin A had higher oestrogenic and lower radiogenic differentiation potential. Their studies demonstrated that lamin A/C played a significant role in the differentiation towards both osteoblast and adipocyte lines by regulating some of the elements of Wnt/β-catenin signaling during early MSCs differentiation, indicating that MSC over expressing Lamina A have higher osteogenic and lower adipogenic differentiation potential. Our results were coincident with Bermeo’s results, because of we found high levels of Lamina A/C by western in MSCs from adult group which we could link to lowest adipogenic potential with the statistically significant (p < 0.05) lowest levels of oil red staining during its directed differentiation (Fig. 2A,B). Also we consider the role of Lamin A like a senescence marker and its relationship whit increase of ROS37 together with increase of Thioredoxin found in our iTRAQ study in adult group could indicate the loss of functionality with age, it might be due to the accumulation of oxidative damage also induce because decrease of SOD2 in this adult group. Stolzing et al.38 demonstrated that age influences impairment of mesenchymal progenitor cells function.
55 modulated proteins found were involved in energetic metabolism process. The decision to exit pluripotency and undergo differentiation is of singular importance for pluripotent cells, including MSCs. The molecular mechanisms for these decisions to differentiate, as well as reversing those decisions during induced pluripotency have focused largely on transcriptomic controls. Easley et al.39 explored the role of translational control for the maintenance of pluripotency and the decisions to differentiate. ATP-citrate synthase is deep linked to pentose phosphate route and its inhibition has been recently linked to a decrease in proliferation rate40. Also it has been reported by Pattapa et cols.41 that MSCs resided under hypoxic conditions which were associated with the inherent metabolism of the cells. However MSCs under normoxia growth conditions derived a significant proportion of ATP from oxidative phosphorylation in addition to glycolysis. The observed increase of LDH in MSCs from adult group (Fig. 4C) could be explained because this group also had the glycolysis increased, on the other way glycolysis decreased in pre-pubertal group. All these results have been supported with our results through pentose phosphate pathway activity because of were significantly decreased G6PDH in pre-pubertal group and significantly increased in adult group (Fig. 4D).
Global protein translation is significantly reduced in hESCs compared to their differentiated progeny. mTOR is a Ser/Thr protein kinase that functions as an ATP and amino acid sensor to balance nutrient availability and cell growth42. mTOR regulates cellular senescence and drives bioenergetic infrastructure12. mTOR restrains proliferation potential of stem cells mediating their self-renewal loss, which is an effect that can be suppressed by mTOR-inhibitors, such as rapamicyn, antagonizing senescence43. mTOR plays an important role in the regulation of hematopoietic stem cell self-renewal in vitro and inhibition of mTOR hyperactivation with rapamycin may represent a novel approach to promote ex vivo expansion and their long-term hematopoietic reconstitution of hematopoietic stem cells44. Our results of mTOR family by western blot analysis indicated that mTOR (Fig. 5A,B) was statistically significant increased in adult group versus the other groups studied it could mean that MSCs from adult group were more senescence than MSCs from younger groups and this result was corroborated by expression of Lamin A/C in adult group in front at no expression of Lamin A in the other groups (Fig. 3B) together with less expression of TMRM (Fig. 5C) in adult group.
We found correlation between inhibition of mTOR and decrease of CD117 and Ki67 which are proliferation markers in the literature45,46 and we wonder if this relationship between mTOR pathway and proliferation was present when the proliferation markers were inhibited by specific reagents. Our results using inhibitors of CD117 and Ki67, imatinib mesylate and JK184 respectively, indicated that mTOR pathway can be modified through modification of proliferation markers, because the MSCs from adult group was treated with the Ki67 inhibitor at two physiological dose which do not affected their viability (Fig. 5D) but the expression of mTOR was modified when the proliferation marker Ki67 and CD117 were diminished (Fig. 5F).
Conclusions
The importance of our study lies in the fact that age from the MSCs source directly influences their differentiation, proliferative and metabolism profiles and also it is the first time where is shown the direct influence of proliferative markers CD117 and Ki67 on activation of mTOR pathway. Summary we affirm that young group of rats has the most proliferative and pluripotent MSCs be able to future functional studies.
Additional Information
How to cite this article: Fafián-Labora, J. et al. Influence of age on rat bone-marrow mesenchymal stem cells potential. Sci. Rep. 5, 16765; doi: 10.1038/srep16765 (2015).
Acknowledgments
The authors wish to thank Mrs. Purificación Filgueira Fernández and Mrs. Noa Goyanes and Mrs. María José Sánchez Dopico for technical assistance. This study was supported by a grant EM2013/011(Xunta de Galicia, Spain, to S.S.-A.) and a grant from Instituto de Salud Carlos III - Ministerio Economía y Competitividad N° Expediente PI11/02799 Unión Europea - Fondo Europeo de Desarrollo Regional (FEDER) “Una manera de hacer Europa”
Footnotes
Author Contributions J.F.-L. realized validation experiments P.F.-P. carried out the isolation of MSCs. S.S. realized the enzymatic experiments. I.F. and N.O. realized the statistical analysis. J.D.T. realized important suggestions which improving the manuscript. J.M. and M.C.A. conceived the study, participated in its design and coordination, and drafted the manuscript. J.F.-L., P.F.-P., J.D.T., N.O., S.S.-A., J.M. and M.C.A. have given final approval of the version to be published.
References
- Murphy M. B., Moncivais K. & Caplan A. I. Mesenchymal stem cells: environmentally responsive therapeutics for regenerative medicine. Experimental & Molecular Medicine. 45, e54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M., Schimmelpfennig C. & Stolzing A. Influence of murine mesenchymal stem cells on proliferation, phenotype, vitality, and cytotoxicity of murine cytokine-induced killer cells in coculture. PLoS One. 9, e88115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. The Effects of Mesenchymal Stem Cells in Craniofacial Tissue Engineering. Current Stem Cell Research & Therapy. 9, 280–289 (2014). [DOI] [PubMed] [Google Scholar]
- Yeh H. Y. et al. Neocartilage formation from mesenchymal stem cells grown in type II collagen-hyaluronan composite scaffolds. Differentiation. 86, 171–183 (2014). [DOI] [PubMed] [Google Scholar]
- Song J. Q. et al. Effect of treadmill exercise timing on repair of full-thickness defects of articular cartilage by bone-derived mesenchymal stem cells: an experimental investigation in rats. PLoS One. 9, e90858 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H. J. et al. Comparative analysis of human mesenchymal stem cells from bone marrow, adipose tissue, and umbilical cord blood as sources of cell therapy. International Journal of Molecular Sciences. 14, 17986–18001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharibi B. et al. Inhibition of Akt/mTOR attenuates age-related changes in mesenchymal stem cells. Stem Cells. 32, 2256–2266 (2014). [DOI] [PubMed] [Google Scholar]
- Tawonsawatruk T. et al. Growth kinetics of rat mesenchymal stem cells from 3 potential sources: bone marrow, periosteum and adipose tissue. Journal of the Medical Association of Thailand. 95, 189–197 (2012). [PubMed] [Google Scholar]
- Geißler S. et al. Functional comparison of chronological and in vitro aging: differential role of the cytoskeleton and mitochondria in mesenchymal stromal cells. PLoS One. 7, e52700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani C. et al. Morphological, molecular and functional differences of adult bone marrow- and adipose-derived stem cells isolated from rats of different ages. Experimental Cell Research. 318, 2034–2048 (2012). [DOI] [PubMed] [Google Scholar]
- Tsai C. C. et al. Hypoxia inhibits senescence and maintains mesenchymal stem cell properties through down-regulation of E2A-p21 by HIF-TWIST. Blood. 117, 459–469 (2011). [DOI] [PubMed] [Google Scholar]
- Han J. et al. Nanog reverses the effects of organismal aging on mesenchymal stem cell proliferation and myogenic differentiation potential. Stem Cells. 30, 2746–2759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N. & Sonenberg N. Upstream and downstream of mTOR. Genes & Devolopment. 18, 1926–1945 (2004). [DOI] [PubMed] [Google Scholar]
- Anisimov V. N. et al. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 10, 4230–4236 (2011). [DOI] [PubMed] [Google Scholar]
- Menendez J. A. et al. mTOR- regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle. 10, 3658–3677 (2011). [DOI] [PubMed] [Google Scholar]
- Mearadji A. et al. Decrease of CD117 expression as possible prognostic marker for recurrence in the resected specimen after imatinib treatment in patients with initially unresectable gastrointestinal stromal tumors: a clinicopathological analysis. Anticancer Drugs. 19, 607–612 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang N. et al. Biodegradable polymeric micelles encapsulated JK184 suppress tumor growth through inhibiting Hedgehog signaling pathway. Nanoscale. 7, 2609–2624 (2015). [DOI] [PubMed] [Google Scholar]
- Karaoz E. et al. Characterization of mesenchymal stem cells from rat bone marrow: ultrastructural properties, differentiation potential and immunophenotypic markers. Histochemistry and Cell Biology. 132, 533–546 (2009). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Matsushime H. et al. D-type cyclin-dependent kinase activity in mammalian cells. Molecular and Cellular Biology. 14, 2066–2076 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research. 39, 561–568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harting M., Jimenez F., Pati S. et al. Immunophenotype characterization of rat mesenchymal stromal cells. Cytotherapy. 10, 243–253 (2008). [DOI] [PubMed] [Google Scholar]
- Mateos J. et al. Proteomic applications in the study of human mesenchymal stem cells. Proteomes. 2, 53–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadaliha M. et al. Quantitative proteomic analysis of human embryonic stem cell differentiation by 8-plex iTRAQ labelling. PLoS One. 7, e38532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen T. & Gerdes J. The Ki-67 protein: from the known and the unknown. The Journal of Cellular Physiology. 182, 311–322 (2000). [DOI] [PubMed] [Google Scholar]
- Blazquez-Martinez A. et al. C-Kit identifies a subpopulation of mesenchymal stem cells in adipose tissue with higher telomerase expression and differentiation potential. Differentiation. 3, 147–160 (2014). [DOI] [PubMed] [Google Scholar]
- Das S. et al. “Over-expression of 60s ribosomal L23a is associated with cellular proliferation in SAG resistant clinical isolates of Leishmania donovani. PLoS Neglected Tropical Diseases. 7, e2527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma-Jonik A. et al. Active heat shock transcription factor 1 supports migration of the melanoma cells via vinculin down-regulation. Cellular Signaling. 27, 394–401 (2015). [DOI] [PubMed] [Google Scholar]
- Piltti J. et al. Rho-kinase inhibitor Y-27632 increases cellular proliferation and migration in human foreskin fibroblast cells. Proteomics. 15, 2953–2965 (2015). [DOI] [PubMed] [Google Scholar]
- Holle A. W. et al. In situ Mechanotransduction via Vinculin regulates stem cell differentiation. Stem Cells. 31, 10.1002/stem.1490. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujalska I. J. et al. A swith in dehydrogenase to reductase activity of 11β-hydroxysteroid dehydrogenase type 1 upon differentiation of human omental adipose stromal cells. Mech Ageing Dev. 124, 747–757 (2013). [Google Scholar]
- Yoon Y. S. et al. Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. Journal of Cellular Physiology. 209, 468–480 (2006). [DOI] [PubMed] [Google Scholar]
- Wu J. et al. TGF-β1 induces senescence of bone marrow mesenchymal stem cells via increase of mitochondrial ROS production. BMC Developmental Biology. 14, 10.1186/1471-213X-14-21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terme J. M. et al. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. The Journal of Biological Chemistry. 286, 35347–35357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne G. A. et al. Nanog Requires BRD4 to Maintain Murine Embryonic Stem Cell Pluripotency and Is Suppressed by Bromodomain Inhibitor JQ1 Together with Lefty1. Stem Cells Development. 24, 879–891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermeo S. et al. Lamin A/C Acts as an Essential Factor in Mesenchymal Stem Cell Differentiation through the Regulation of the Dynamics of the Wnt/β-Catenin Pathway. Journal Cell Biochemistry. 4, 10.1002/jcb.25185. (2015). [DOI] [PubMed] [Google Scholar]
- Mateos J., De la Fuente A., Lesende-Rodriguez I. et al. Lamin A deregulation in human mesenchymal stem cells promotes an impairment in their chondrogenic potential and imbalance in their response to oxidative stress. Stem Cell Res. 11, 1137–1148 (2013). [DOI] [PubMed] [Google Scholar]
- Stolzing A. & Scutt A. Age-related impairment of mesenchymal progenitor cell function. Aging Cell. 5, 213–224 (2006). [DOI] [PubMed] [Google Scholar]
- Easley C. A. et al. mTOR- mediated activation of p70 S6K induces differentiation of pluripotent human embryonic stem cells. Cell Reprogramming. 12, 263–273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. et al. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Molecular Cell. 51, 506–518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pttappa G. et al. The metabolism of human stem cells during proliferation and differentiation. J Cell Physiol. 226, 2562–2570 (2011). [DOI] [PubMed] [Google Scholar]
- Sabers C. J. et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. The Journal of Biological Chemistry. 270, 815–822 (1995). [DOI] [PubMed] [Google Scholar]
- Martins I., Galluzzi L. & Hormesis Kroemer G. cell death and aging. Aging. 3, 821–828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. et al. Rapamycin enhances long-term hematopoietic reconstitution of ex vivo expanded mouse hematopoietic stem cells by inhibiting senescence. Transplantation. 97, 20–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillenburg C. S., Martins M. D., Meurer L., Castilho R. M. & Squarize C. H. Keratoacanthoma of the Lip: Activation of the mTOR Pathway, Tumor Suppressor Proteins, and Tumor Senescence. Medicine. 94, e1552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A. et al. Differential Reponses of Hematopoietic Stem and Progenitor Cells to mTOR Inhibition. Stem Cells Int. 561404, 10.1155/2015/561404. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]