Fig. 4.
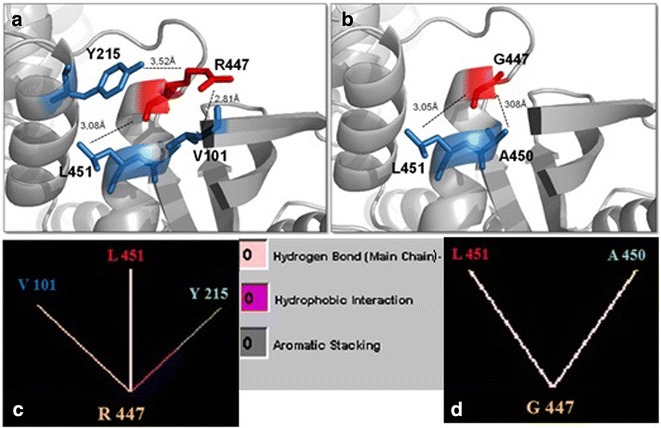
Comparison of normal and mutant glucokinase models at residue 447. a, b Pymol structures for normal arginine and mutant glycine, respectively. Each residue is denoted in red and surrounding residues in blue. Dashed lines represent the distance in Ångström (Å) for internal contacts. c, d Graphic results for residue interactions obtained in the STING Millennium analysis. a, c The native Arg447 makes two hydrogen bonds with Val101 and with Leu451, and interacts with Tyr215 through aromatic stacking and hydrophobic interactions. b, d The mutant Gly447 looses both Val101 and Tyr215 interactions, creates a new hydrogen bond with Ala450, and maintains the hydrogen bond with Leu451. Between c and d, color legend for internal interactions provided by BlueStar STING software
