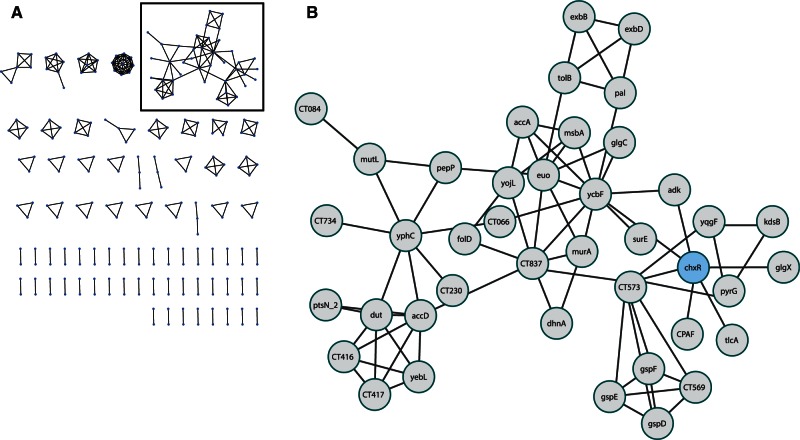
Fig. 7.
Phylum-wide conservation of predicted coregulations in the Chlamydiae. The consensus network was created by comparison of individual organism’s predicted coregulation networks (n = 6). An edge was kept if it was present in five or more networks. The 240 genes present in this network (A) mainly represent conserved operons, with the exception of a putative regulon of 41 genes (B) that mainly have function in exploitation of the intracellular niche. The activator of this subnetwork may be ChxR, which is indicated. Notably, the virulence genes detected in individual organism’s networks are absent in this consensus network.