Figure 4.
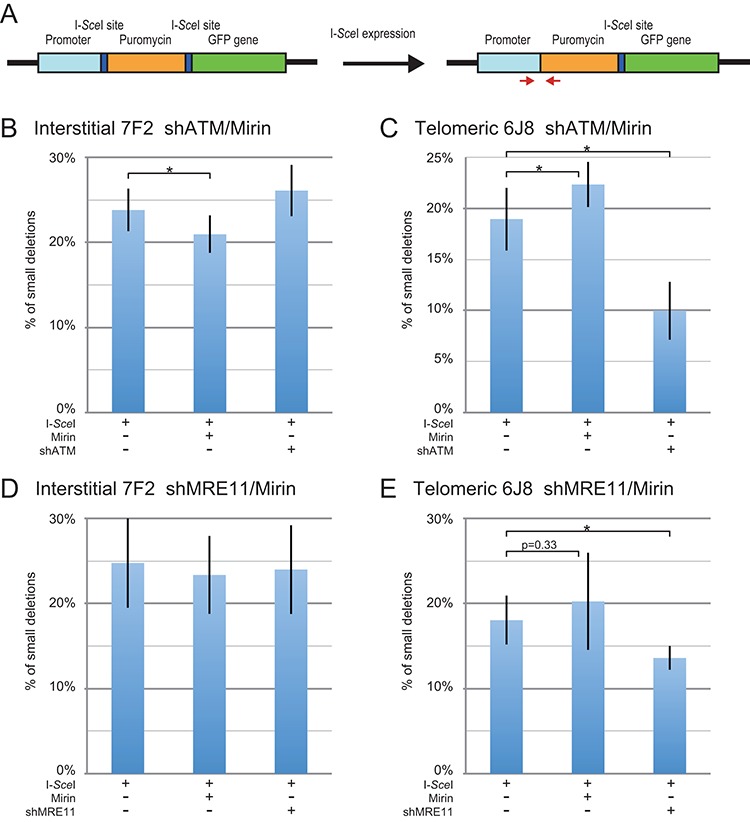
The effect of inhibition of MRE11 and ATM on small deletions at interstitial and subtelomeric DSBs. (A) Cell clones containing the pEJ5-GFP plasmid integrated at an interstitial (EDS-7F2) or telomeric (EDS-6J8) site were used for analysis of small deletions. Small deletions were determined by first amplifying a PCR product that spans one of the I-SceI endonuclease recognition sites from genomic DNA isolated from the pooled population of cells expressing I-SceI endonuclease. The PCR product was then digested with I-SceI endonuclease to determine the frequency of cells in the population with small deletions at the I-SceI-induced DSB, as shown by the fraction of PCR product that is not cut with I-SceI endonuclease. The frequency of small deletions at the I-SceI-induced DSB was determined for clone EDS-7F2 (B, D) and clone EDS-6J8 (C, E) following infection with the pQCXIH-ISceI retrovirus vector and selection with hygromycin for 14 days for EDS-7F2 and 15 days for EDS-6J8. Small deletions were analyzed following (B, C) treatment with Mirin or knockdown of ATM (shATM), or (D, E) treatment with Mirin or knockdown of MRE11 (shMRE11). Control cultures for knockdown of ATM or MRE11 were treated with shRNA-mediated knockdown of luciferase, while control cultures for Mirin were treated with DMSO. The values shown in the graph represent the average of more than three independent experiments, each done in triplicate. Error bars represent the standard deviation of more than three separate experiments. Statistical significance for comparisons between the indicated values (horizontal lines) was determined using the two-tailed Student's t-test, and an asterisk indicates statistically significant values of 0.05 or less.
