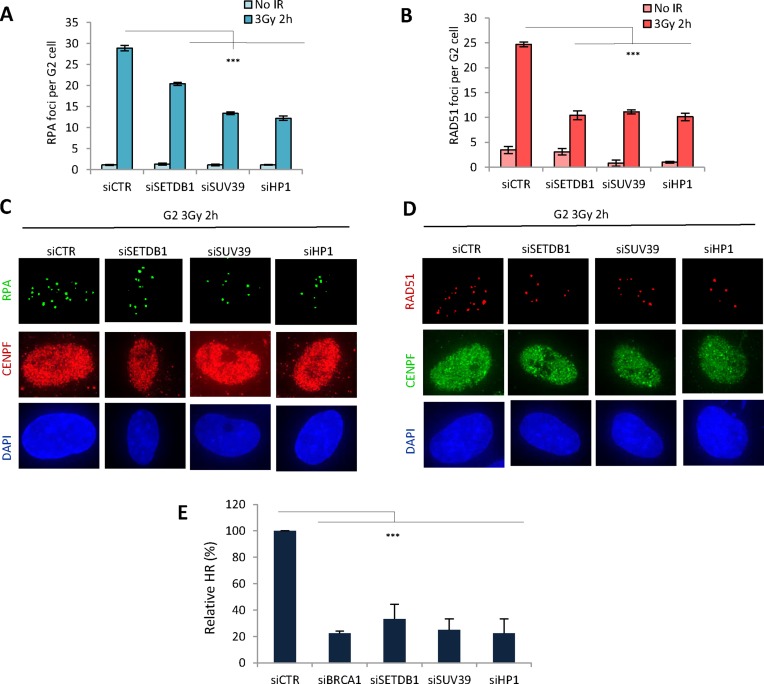
Figure 2.
SETDB1, SUV39 and HP1 are required for the formation of RPA and RAD51 foci and for HR. (A and B) 1BR3 hTERT cells were transfected with siRNAs targeting control (CTR), SETDB1, SUV39, HP1 and BRCA1, irradiated with 3 Gy and 2 h post IR, (A) RPA and (B) RAD51 foci were quantified. RPA and RAD51 analysis at additional time points is shown in Supplementary Figure S1. Asterisks denote statistically significant differences between control (CTR) and SETDB1, SUV39, HP1 knockdown cells. (P < 0.001;t-test). (C and D) Representative images from the above experiments: (C) RPA and CENPF (D) RAD51 and CENPF as indicated. E. U2OS DR-GFP cells were transfected with siRNAs targeting control (CTR), BRCA1, SETDB1, SUV39 or HP1 and 24 h later were re-transfected with pCBASceI. A GFP signal is generated following intrachromosomal gene conversion following I-SceI-induced DSB formation. The percentage of GFP+ cells was measured by FACS 48 h after transfection with I-Sce1. GFP+ cells were normalized to the percentage of EGFP positive cells and the level of the S/G2 population. All samples had a similar fraction of G2 phase cells and expression of I-Sce1 was shown to be similar by Western blotting using an HA-tag on I-Sce1 (Supplementary Figure S2A, B). Asterisks denote statistically significant differences (P<0.001; t-test). Results represent the mean ± S.E.M of 3 experiments.