Figure 5.
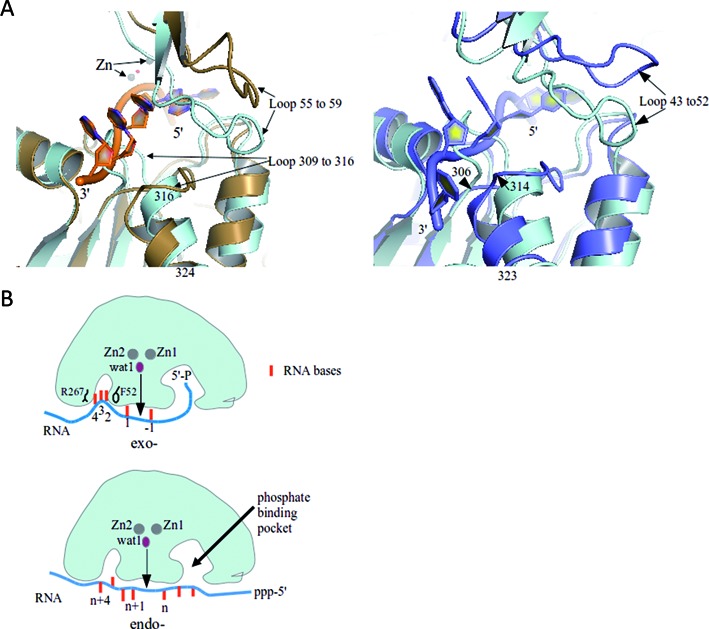
Conformational adjustments in RNase J with RNA binding and the substrate path for the exo- and endo-cleavage modes. (A) The left panel presents a structural overlay of the RNA-bound S. coelicolor RNase J (sand) with the apo-form of Thermus thermophilus RNase J (cyan) (PDB code: 3BK1). Loop movements may accommodate the RNA molecule (backbone in orange and base shown as filled rings). The panel on the right compares the T. thermophilus RNase J apo-form (cyan) and RNA-bound form (blue), where the RNA bears 2′-O-methyl modifications. (B) Proposed substrate interactions for exo- and endo- cleavage modes. Sandwiching of the bases between R267 and F52 and 5′ end sensing are proposed to favor the exo- mode, perhaps through a ‘spring-loaded’ mechanism. In the endo mode, the 5′ binding pocket is potentially left unoccupied. Endo cleavages may involve tetramer dissociation and re-association on the substrate and potentially some translocation along tunnels (see Supplementary Figure S5E for possible pathways for the tunnels).
