Figure 4.
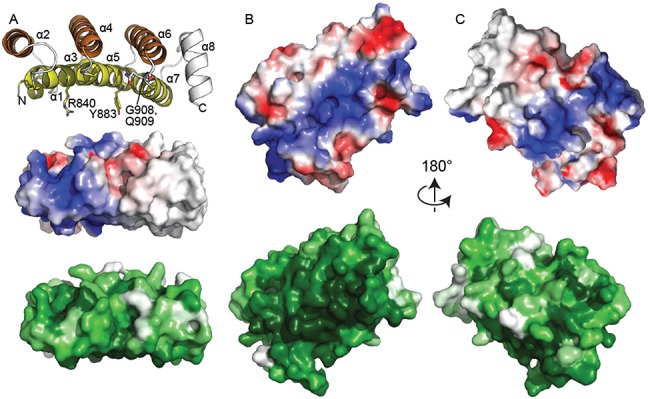
The concave surface of DM15 is conserved and positively charged. (A) Edge-on views of DM15 reveal that it assumes a curved structure. Top, cartoon representation and coloured as in Figure 2A. Amino acids tested for their roles in binding the RPS6 5′TOP oligonucleotide are shown as sticks. Middle, surface electrostatic representation as calculated by PyMOL (blue, electrostatically positive; red, electrostatically negative). Bottom, surface representation coloured by conservation as calculated by Consurf (42) using alignment of DM15 sequences from Supplementary Figure S4 (white, less conserved; dark green, more conserved). (B) View of the concave DM15 surface, coloured as in (A). (C) View of the convex DM15 surface, coloured as in (A). The view in panel (B) is related to that in panel (C) by a 180° rotation about the x-axis.
