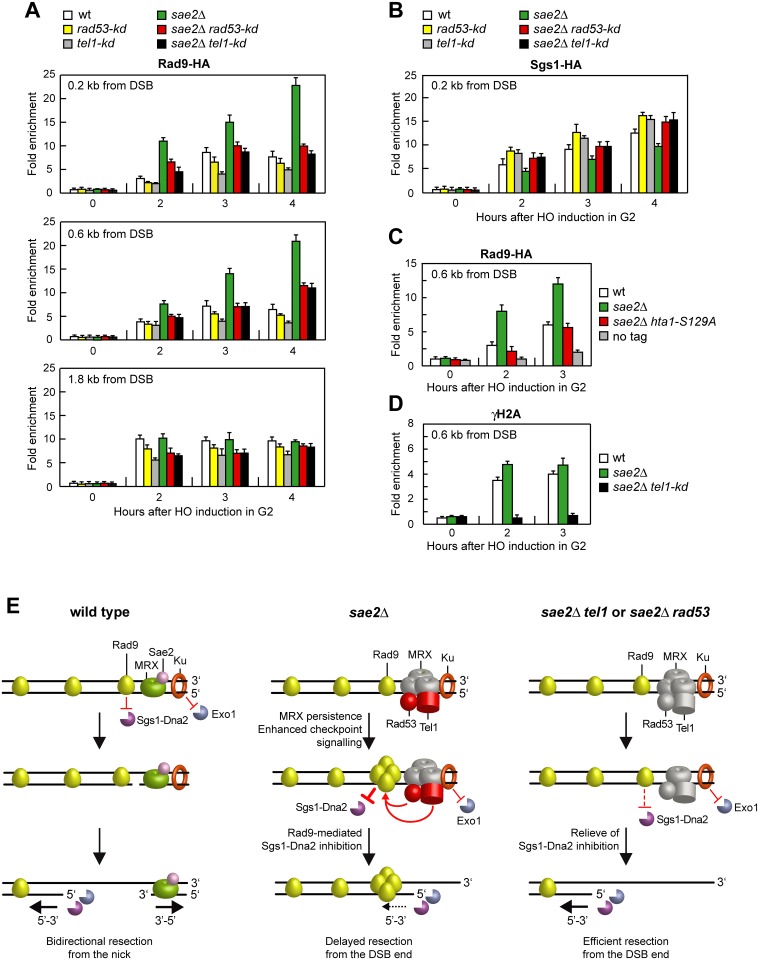
Fig 10. Rad53-kd and Tel1-kd prevent Rad9 association at DSBs.
(A) Exponentially growing YEPR cell cultures of JKM139 derivative strains were arrested in G2 with nocodazole and transferred to YEPRG in the presence of nocodazole. Recruitment of Rad9-HA at the indicated distance from the HO-cut was determined by ChIP and qPCR. In all diagrams, the ChIP signals were normalized for each time point to the amount of the corresponding input signal. The mean values are represented with error bars denoting s.d. (n = 3). (B) As in (A), but showing Sgs1-HA binding. (C) As in (A). All strains carried also the deletion of HTA2 gene. (D) As in (A), but showing γH2A binding. (E) Model for the role of Sae2 at DSBs. (Left) Sae2 activates the Mre11 endonuclease activity to incise the 5’ strand. Generation of the nick allows bidirectional processing by Exo1/Sgs1-Dna2 in the 5’-3’ direction from the nick and MRX in the 3’ to 5’ direction toward the DSB ends. Ku and Rad9 inhibit DSB resection by limiting Exo1 and Sgs1-Dna2, respectively. (Middle) The absence of Sae2 impairs the MRX nuclease activity (non functional MRX nuclease is in grey). As a consequence, the endonucleolytic cleavage of the 5’ strand does not occur and resection is carried out by Exo1 and Dna2-Sgs1 that degrade the 5’ strands from the DSB ends. Impairment of Mre11 nuclease activity also causes increased MRX association at the DSB, which leads to enhanced Tel1-dependent Rad53 activation. Tel1 and Rad53 activities limit DSB resection from the DSB end (dashed arrow) by increasing the amount of DSB-bound Rad9, which inhibits Sgs1-Dna2 recruitment at DSBs. (Right) Impairments of Tel1 or Rad53 activity (non functional Tel1 and Rad53 are in grey) restore efficient resection in sae2Δ cells by relieving Rad9-mediated inhibition of Sgs1-Dna2. Restored DSB resection by Sgs1-Dna2 also reduces MRX persistence at the DSB.