Figure 1. Differential gene expression profile in response to DCLK1 overexpression in the presence or absence of HCV.
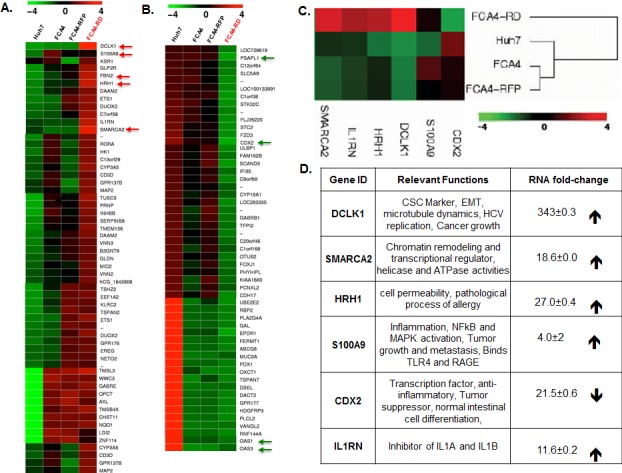
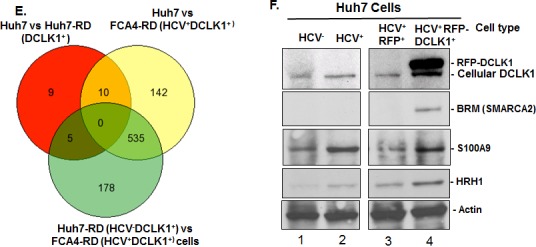
The total RNAs isolated from Huh7 hepatoma cells expressing HCV subgenomic replicon (FCA4, HCV+) and FCA4 cells overexpressing recombinant RFP-DCLK1 (FCA4-RD, HCV+DCLK1+) were subjected to transcriptome analysis by RNASeq method. The data were analyzed using GeneSifter software at 10-fold change in the expression level and at least 10 reads (Quality). The results are derived from two independent biological and sequencing replicates. The FCA4-RFP (FCA4 expressing GFP), Huh7 and FAC4 were used as controls for the comparison of FCA4-RD transcriptome. A. and B. Heat-maps for differential gene expression in FCA4-RD cells. C. Cluster analysis for the selected genes. D. The reported gene functions and fold-changes in FCA4-RD RNAs with standard deviations as compare to the controls cell lines. E. Venn diagram showing total number of common and differentially regulated genes as indicated. F. Validation of three differentially regulated gene products by Western blot using cell lysates (30 μg). HCV and DCLK1 co-expression induces BRM/SMARCA2 expression and inflammatory molecules (S100A9 and HRH1).
