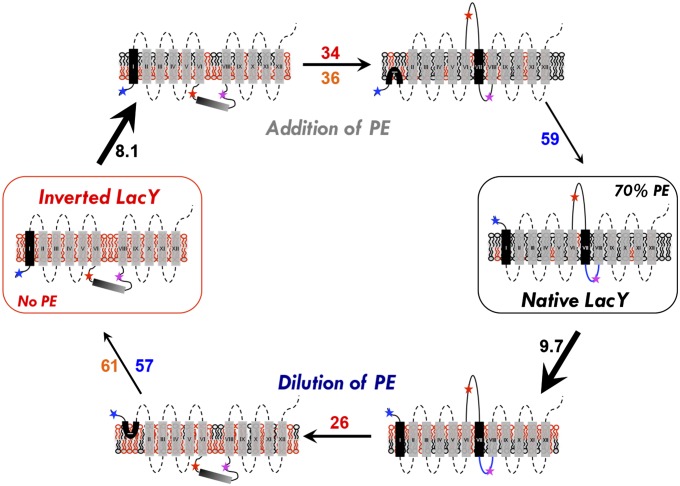
Fig. 5.
Schematic model of the events occurring during lipid-induced topological switching of LacY in proteoliposomes. LacY topology is shown when assembled in proteoliposomes lacking PE (inverted LacY, Left) and in proteoliposomes containing WT amounts of PE (Native LacY, Right), before and during lipid exchange. Stars indicate positions of diagnostic Trp replacements used to determine topological orientation of EMDs (blue, red, and magenta correspond to NT, C6, and P7, respectively). The blue P7 loop indicates proper folding of the P7 epitope. Membrane phospholipids are depicted as follows: black, PE; red, anionic lipids (PG and CL). The time constant value (s−1) of each event is indicated as follows: black, lipid exchange; red, C6 flipping; blue, NT flipping; orange, lipid flipping. In both addition and dilution of PE (or PC), lipid exchange occurs first and is followed by protein topological rearrangement and transbilayer movement of lipid. During lipid exchange, lipids from the outer leaflet of proteoliposomes are exchanged and become enriched with PE (addition of PE) or PG/CL (dilution of PE), leading to transient generation of lipid asymmetry. Protein flipping occurs subsequent to lipid exchange, with C6 EMD flipping being more sensitive to PE (or PC) content than the NT EMD. P7 EMD folding/unfolding is PE-dependent and linked to the topological switch. The rate of C6 EMD flipping is faster during PE dilution, which may be due to specific interaction of LacY with PE. Asymmetrical distribution of lipids across the lipid bilayer may accelerate the rate of protein flipping.