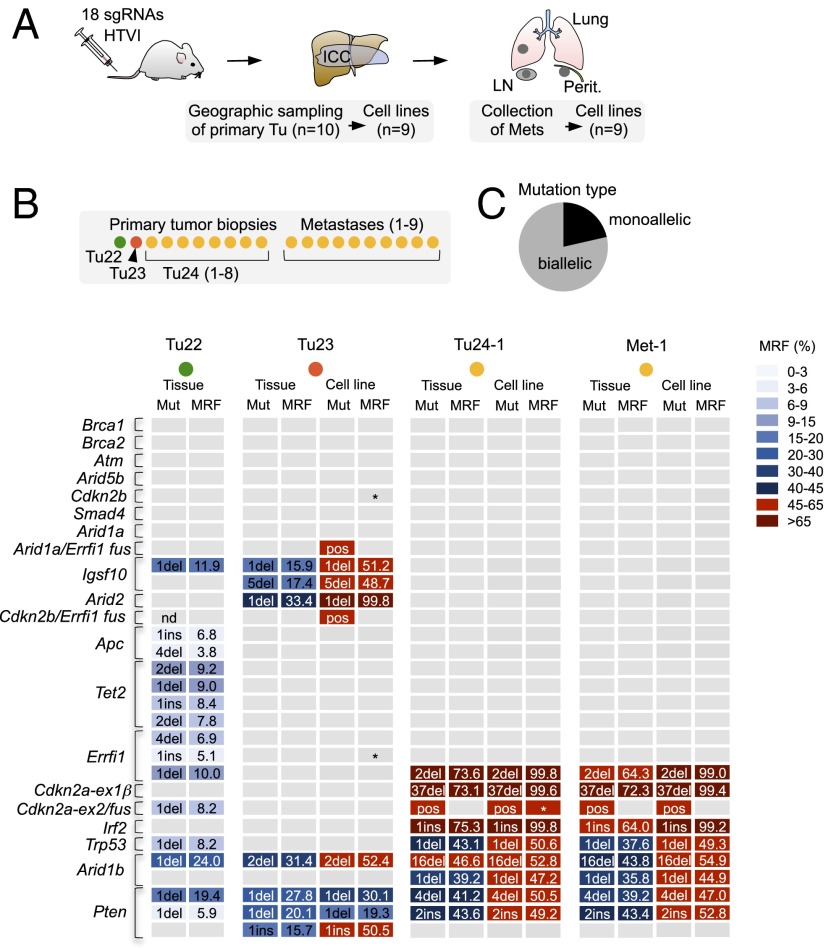
Fig. 4.
Allelic frequencies of target site mutations and phylogenetic tracking of CRISPR/Cas9-induced metastatic ICC. (A) A 2-cm tumor mass (ICC) and numerous metastases (Mets) in lungs, lymph nodes (LN), and peritoneum in a mouse 20 wk after HTVI of 18 sgRNAs. (B) CRISPR/Cas9 target sites underwent NGS in a total of 35 tumor/metastasis tissues and cell lines. Indel patterns revealed three independent primary tumors. All metastases originate from Tu24. Frame-shift causing indels with a cumulative mutant read frequency (MRF) >4% per target site are shown for representative samples. Note that MRFs are underestimated in cancer tissue (because of healthy stromal components) but are accurately reflected in cell lines. Tumors with indicated fusion products are marked as positive (pos). Asterisks indicate a lack of WT sequence. (C) Allelic frequency of mutations in cell lines of Tu23 and Tu24 as determined by a combined quantitative analysis of indel frequencies, presence/absence of large fusions, and the presence/absence of WT reads.